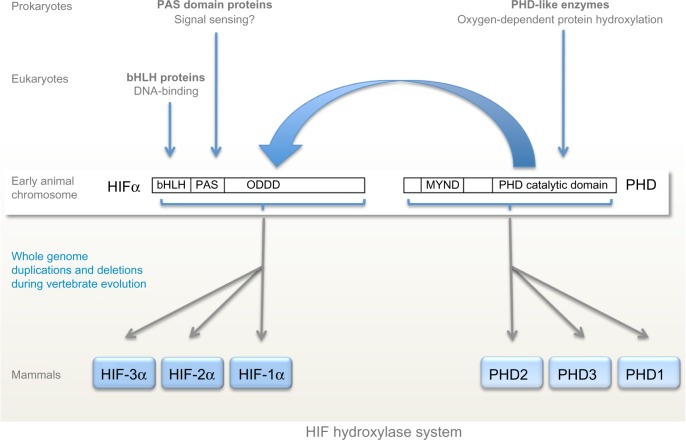
Figure 2.
Evolution of the hypoxia-inducible factor (HIF) hydroxylase system.
Notes: The major components of the HIF hydroxylase system are widely represented across both animal and non-animal species. PAS domains and PHD-like enzymes are found in both prokaryotes and eukaryotes whilst bHLH domains are found in all eukaryotes. The HIF hydroxylase system appears to have emerged with the evolution of early animals, coinciding with the convergence of bHLH and PAS domains to form bHLH/PAS-containing proteins including HIF-α. Despite that coincidence, it is sequences lying distal to the bHLH/PAS domains (ODDD) that are sensitive to oxygen-dependent post-translational hydroxylation (large blue arrow). In Trichoplax adhaerens, considered to be the “basal” animal, the HIF hydroxylase system consists of a single PHD enzyme and HIF-α, whose genes lie on the same chromosome. Whole genome duplications and deletions during vertebrate evolution result in multiple copies of both HIF-α and PHD in mammals.
Abbreviations: PAS, PER-AHR/ARNT-SIM; PER, period circadian protein; AHR, aryl-hydrocarbon receptor; ARNT, aryl-hydrocarbon-receptor nuclear translocator; SIM, single-minded protein; PHD, prolyl hydroxylase domain; bHLH, basic-helix-loop-helix; ODDD, oxygen-dependent degradation domain; MYND, myeloid, Nervy and DEAF-1.