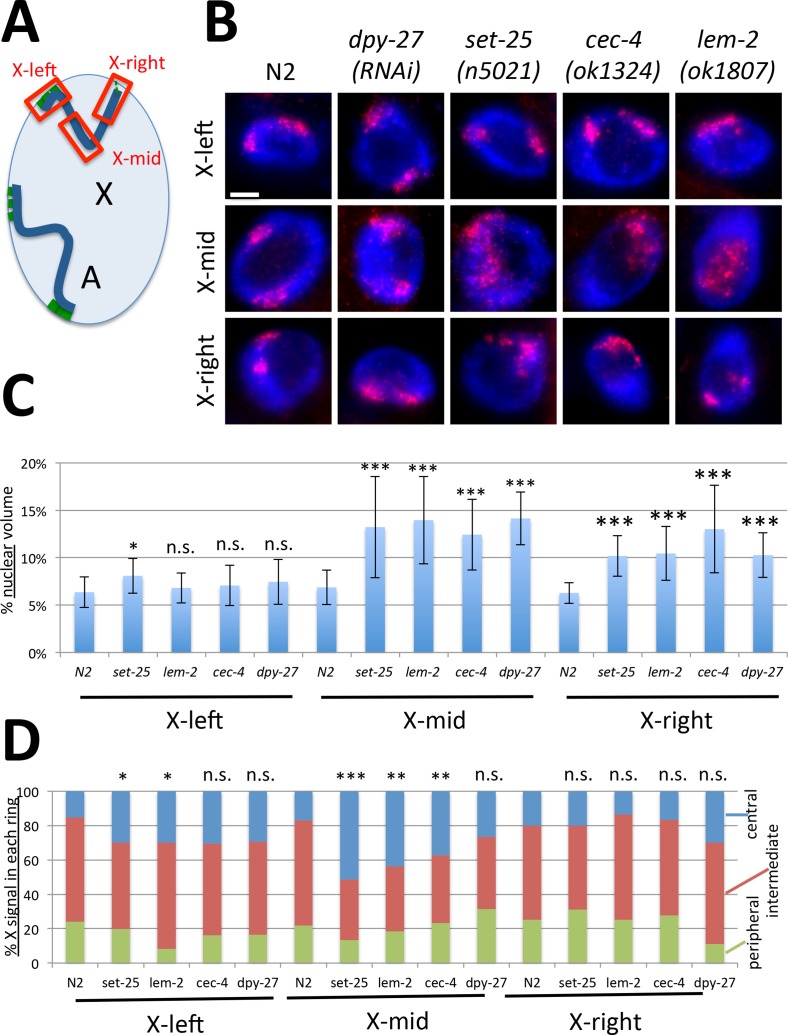
Fig 4. The middle region of the X chromosome is most affected in the absence of heterochromatic tethers.
(A) Autosomes are anchored to the nuclear lamina at both chromosome arms (anchors shown in green), while the X chromosome only has a significant anchored domain at the left end. Probes used in FISH analysis are indicated in red. Each probe covered an approximately 3–4 Mb genomic region. (B) Representative images of X-left, X-mid, X-right FISH analysis in each genotype. The mid-X region appears most decondensed and most centrally located in mutants. Scale bar, 5 μm. (C) Quantification of volumes occupied by the indicated FISH probes, normalized to nuclear size (n = 12 nuclei). Error bars indicate standard deviation. The greatest degree of decondensation in mutants is observed for the mid-X probe. (D) Three zone assay for each probe (n = 12 nuclei). The greatest degree of central relocation is observed for the mid-X probe. Asterisks indicate statistical analysis of mutant to wild type comparisons of volumes in (C) and centrally located portion of the X in (D) using Student's t-test. n.s. = p>0.05, * = p<0.05, ** = p<0.01, *** = p<0.001. See S1 Table for statistical data.