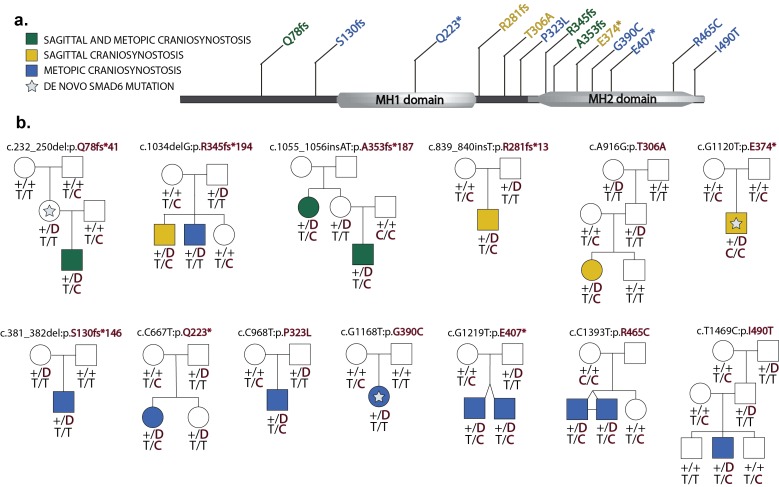
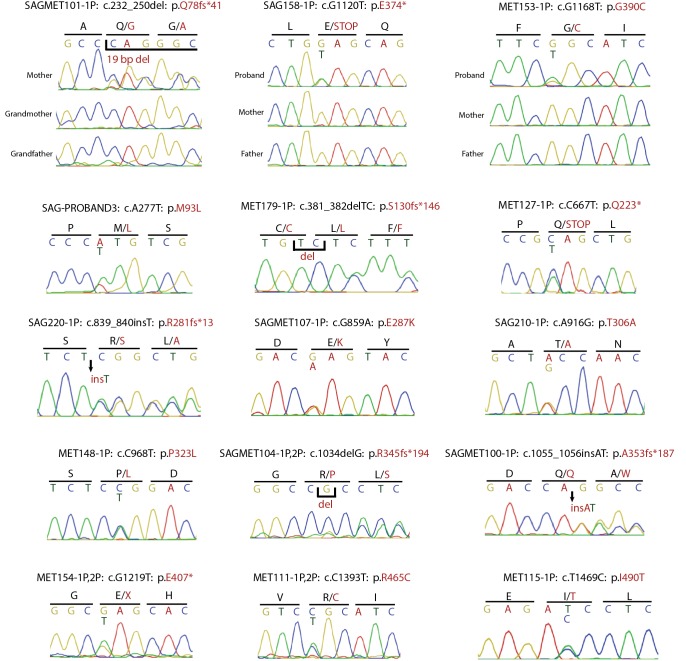
Figure 2. Segregation of SMAD6 mutations and BMP2 SNP genotypes in pedigrees with midline craniosynostosis.
(a) Domain structure of SMAD6 showing location of the MH1 and MH2 domains. The MH1 domain mediates DNA binding and negatively regulates the functions of the MH2 domain, while the MH2 domain is responsible for transactivation and mediates phosphorylation-triggered heteromeric assembly with receptor SMADs. De novo or rare damaging mutations identified in craniosynostosis probands are indicated. Color of text denotes suture(s) showing premature closure. (b) Pedigrees harboring de novo (denoted by stars within pedigree symbols) or rare transmitted variants in SMAD6. Filled and unfilled symbols denote individuals with and without craniosynostosis, respectively. The SMAD6 mutation identified in each kindred is noted above each pedigree. Below each symbol, genotypes are shown first for SMAD6 (with 'D' denoting the damaging allele) and for rs1884302 risk locus downstream of BMP2, (with 'T' conferring protection from and 'C' conferring increased risk of craniosynostosis). All 17 subjects with craniosynostosis have SMAD6 mutations, and 14/17 have also inherited the risk allele at rs1884302, whereas only 3 of 16 SMAD6 mutation carriers without the rs1884302 risk allele have craniosynostosis.
DOI: http://dx.doi.org/10.7554/eLife.20125.006
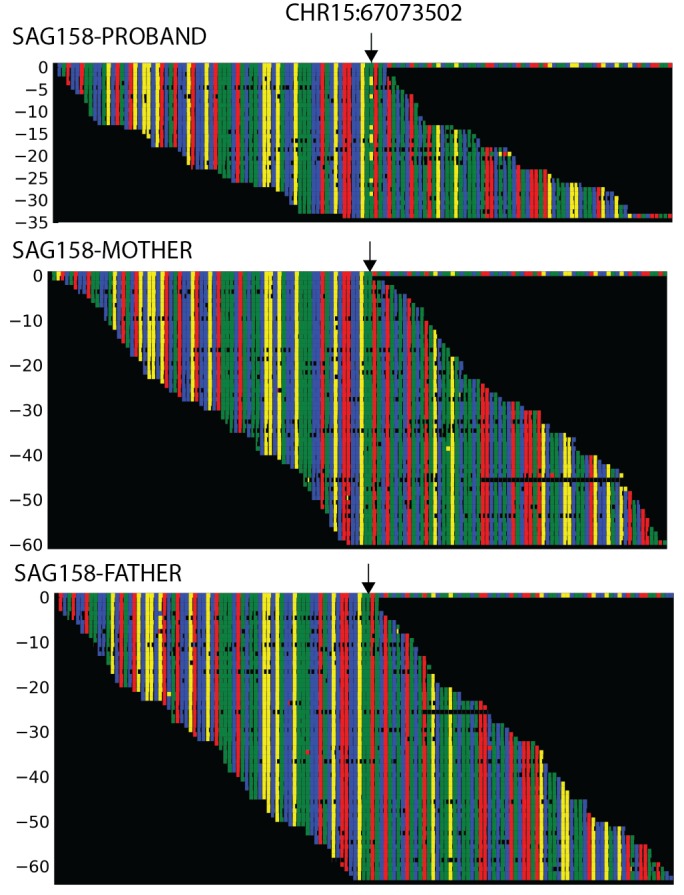
Figure 2—figure supplement 1. Plots of independent Illumina sequencing reads in a parent-offspring trio showing de novo SMAD6 mutation.