Figure 1. GLP-1Notch signaling promotes reprograming of germ cells.
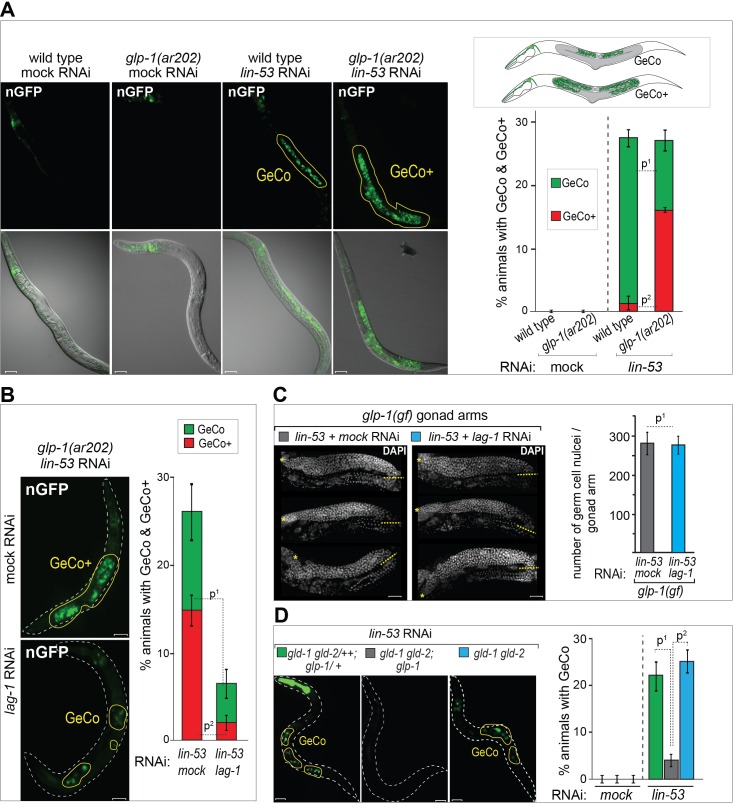
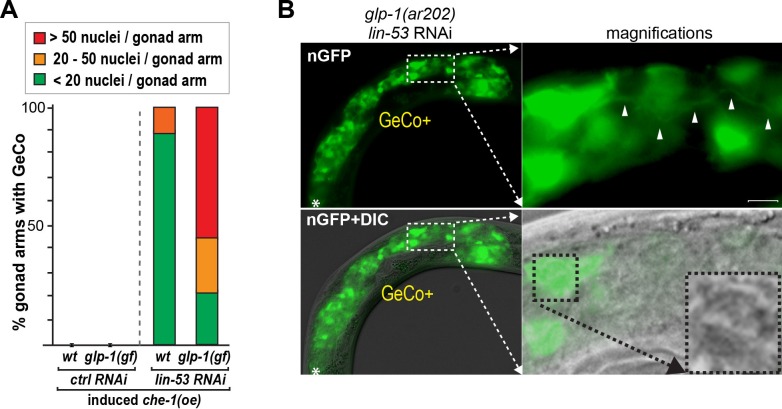
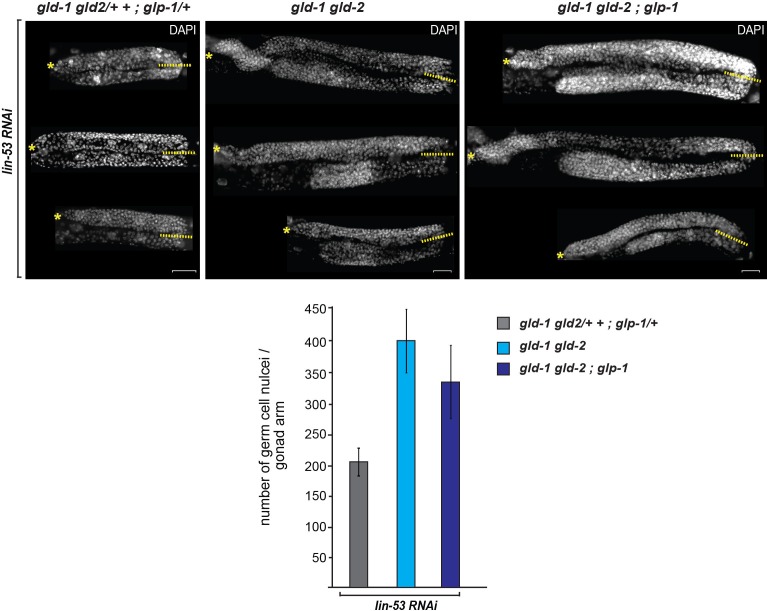
(A) GLP-1Notch enhances germ cell conversion (GeCo) into neuronal-like cells. Left: Fluorescent (top) and combined fluorescent/differential interference contrast (DIC) micrographs (bottom) of adult animals. All animals ectopically expressed the pro-neuronal transcription factor CHE-1 from a heat-shock promoter. glp-1(ar202) is a temperature-sensitive gain-of-function allele of the Notch receptor. Animals were subjected to either mock (control) or lin-53 RNAi. Reprogrammed cells expressed a GFP reporter driven from the neuronal gcy-5 promoter (here an in other figures nGFP) and are outlined here and elsewhere in yellow. Any signal outside the outlined region comes from somatic tissues. GeCo+ indicates animals that displayed a strongly enhanced GeCo phenotype. Scale bars = 10 μm. The cartoons depicting the GeCo and GeCo+ phenotypes are on the top right. The gonads are shaded in grey and GFP-positive converted germ cells are green. Fractions of animals displaying GeCo and GeCo+ are indicated below. At least 250 animals were quantified per condition. P-values were calculated using Student's t-test: p1<0,0001; p2=0,0006. Error bars represent SEM. (B) The transcriptional effector of the GLP-1Notch signaling pathway, LAG-1, is required for the GLP-1Notch–mediated enhancement of GeCo. Left: Fluorescent micrographs of adults expressing CHE-1–induced nGFP as explained above. GeCo is diminished upon the depletion of LAG-1. White dashed lines outline the animal body. Scale bars = 10 μm. Right: The corresponding quantifications. At least 400 animals were quantified per condition. P-values were calculated using Student's t-test: p1<0,0001; p2=0,0018. Error bars represent SEM. (C) GLP-1Notch signaling enhances GeCo independently from germ cell proliferation. Shown are DAPI-stained gonads of glp-1(ar202) animals, expressing CHE-1–induced nGFP, treated with either mock or lin-53 RNAi. Germ cells were counted from the DTC (yellow asterisk) to the turn of the gonad arm (dashed yellow line). 15 gonad arms per condition were counted. Scale bars = 10 μm. Quantifications are on the right. While greatly inhibiting GeCo, lag-1 RNAi did not change the number of germ cells. P-values were calculated using Student's t-test: p1=0,89. Error bars represent SEM. (D) GLP-1Notch enhances GeCo independently from proliferation. Left: Fluorescent micrographs of adults (with indicated genotypes), expressing CHE-1–induced nGFP. The first panel on the left shows a control, heterozygous (wild-type) gld-1 gld-2/++; glp-1/+ animal. The other panels show the homozygous gld-1(q497) gld-2(q485) mutants, carrying either a loss-of-function (q175, center) or a wild-type (right) allele of glp-1. Despite proliferating, germ cells in the gld-1 gld-2; glp-1 gonads have lost the ability to undergo GeCo. Scale bars = 10 μm. Right: the corresponding quantifications. At least 250 animals were quantified per condition. P-values were calculated using Student's t-test: p1=0,0478; p2=0,0201. Error bars represent SEM.
DOI: http://dx.doi.org/10.7554/eLife.15477.003