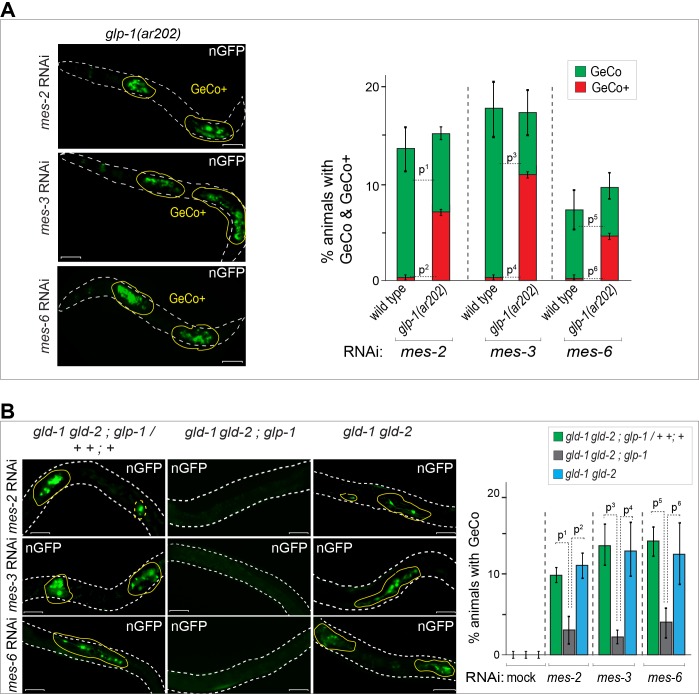
Figure 3. GLP-1Notch enhances reprograming upon the depletion of PRC2.
(A) Notch and PRC2 genetically interact in GeCo. Left: Fluorescent micrographs of glp-1(ar202) gain-of-function mutants expressing CHE-1–induced neuronal GFP. The animals were subjected to control RNAi or RNAi against PRC2 components (MES-2, 3, and 6), as indicated. Increased GLP-1Notch signaling enhanced the GeCo+ phenotype upon PRC2 depletion. Control RNAi (mock) for each genetic background did not result in any GeCo (images not shown – see quantification). Right: The corresponding quantifications. P-values were calculated using Student's t-test: p1=0,0006; p2<0,0001; p3=0,0536; p4=0,0001; p5=0,4035; p6=0,0003. At least 200 animals were scored per condition. Error bars represent SEM. (B) GLP-1Notch is required for GeCo in PRC2-depleted animals independently from proliferation. Left: Fluorescent micrographs of adults expressing CHE-1–induced nGFP, with the genotypes indicated above the panels. The animals were subjected to RNAi as indicated on the left. The first column shows heterozygous, the other two homozygous animals carrying the loss of function alleles gld-1(q497), gld-2(q485) and, in the central panels, glp-1(q175). The animals were subjected to control RNAi or RNAi against PRC2 components (MES-2, 3, and 6). In the absence of GLP-1Notch, depletion of PRC2 components did not induce GeCo. Scale bars = 10 μm. Right: The corresponding quantifications. P-values were calculated using Student's t-test: p1<0,0456; p2=0,0337; p3=0,0070; p4=0,0637; p5=0,0080; p6=0,1259. At least 70 animals were scored per condition. Error bars represent SEM.
DOI: http://dx.doi.org/10.7554/eLife.15477.014