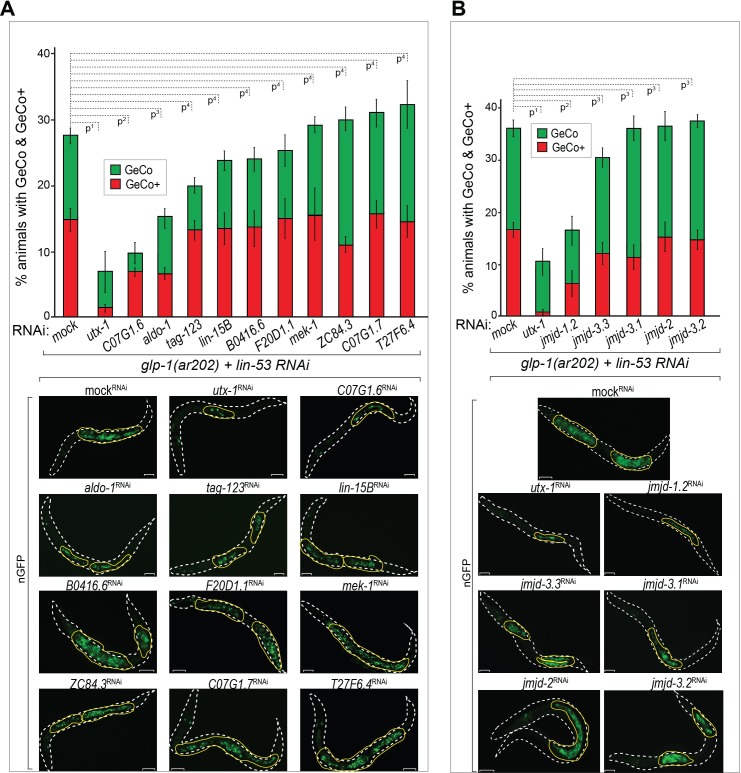
Figure 4. The H3K27 demethylase UTX-1 is required for GeCo enhancement.
(A) UTX-1 is critical for GeCo enhancement. Candidate Notch-activated genes, selected from Supplementary file 2 with available RNAi clones, were assayed for a role in GeCo in glp-1(ar202) animals, expressing CHE-1–induced nGFP and treated with lin-53 RNAi. While the additional depletion of utx-1 had the strongest impact on GeCo+ and GeCo, the depletion of C07G1.6 and aldo-1 had a weaker effect. Representative fluorescence micrographs are below the quantification chart. White dashed line outline the animal body, yellow lines outline gonadal areas with GeCo. P-values for GeCo+ were calculated using Student's t-test: p1=0,000013; p2=0,026; p3=0,021; p4>0,1. At least 250 animals were scored per condition. Error bars represent SEM. nGFP = gcy-5::gfp. Scale bars = 10 μm. (B) As in A, but RNAi was performed against jmjd-1.2 (H3K9/27me2 demethylase); jmjd-3.1, jmjd-3.2, and jmjd-3.3, (H3K27me2/3 demethylases); and jmjd-2 (H3K9/36 demethylase). Only RNAi against jmjd-1.2 suppresses GeCo+, though to a lesser degree compared to utx-1 RNAi. Representative fluorescence micrographs are below the quantification chart. P-values for GeCo+ were calculated using Student's t-test: p1=0,0042; p2=0,035; p3>0,2. At least 190 animals were scored per condition. Error bars represent SEM. Scale bars = 10 μm.
DOI: http://dx.doi.org/10.7554/eLife.15477.016