Figure 2. Identification of CRKL as a downstream signaling molecule of EML4-ALK.
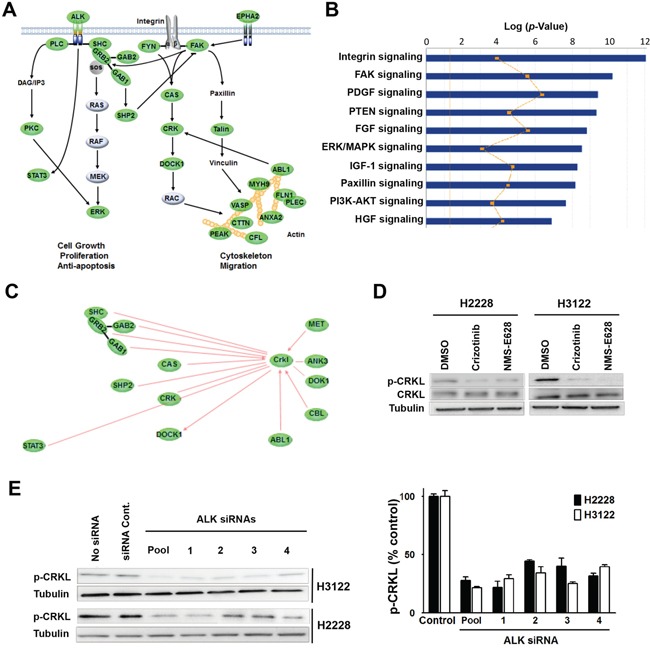
A. Regulatory network sensitive to ALK inhibitors in H3122 and H2228 cells revealed by phosphotyrosine peptide mapping. Core signaling proteins inhibited by ALK inhibitors in H3122 and H2228 cells are shown in green. The proteins with ≥2-fold decrease of phosphorylation (at least one tyrosine residue) 1 hr after treatment are presented. B. IPA analysis of tyrosine-phosphorylated proteins with differential signaling pathways in H3122 and H2228 cells treated with ALK inhibitor. The yellow line indicates the fraction associated with each pathway of genes that were expressed in each cell line. C. Networking of CRKL with various signaling molecules detected in the phosphotyrosine peptide mapping study. Core signaling molecules inhibited by ALK inhibitors in H3122 and H2228 cells are shown in green. D. Validation of decreased CRKL phosphorylation identified by mass spectrometry. Lysates from H2228 and H3122 cells treated with/without crizotinib (900nM) or NMS-E628 (300nM) for 1 hr were subjected to Western blot probed with anti-p-CRKL (Y207) antibody. As controls, total CRKL and Tubulin were also detected. E. Effect of ALK siRNA knockdown (20 nM for 72 h) on CRKL phosphorylation. Western blot analyses were performed on the lysates from H3122 and H2228 cells treated with ALK siRNAs (four individual siRNA or their smartpool at 20 nM for 72 h) to determine p-CRKL (Y207) level. The graph shows the quantification of p-CRKL levels for each treatment (Student's t-test: p-value < 0.05).
