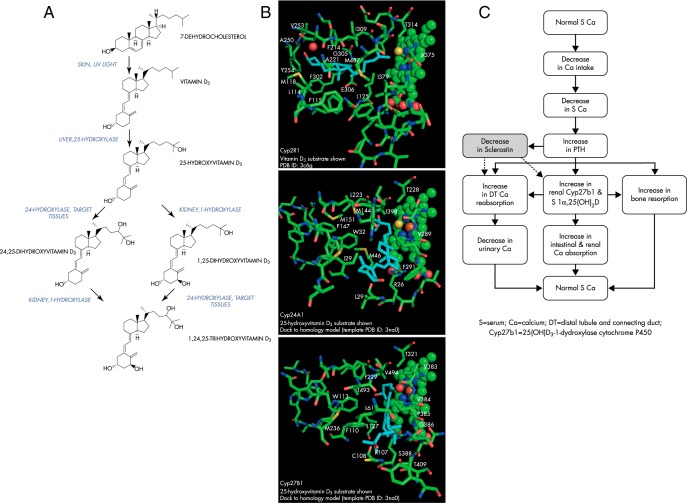
Figure 1.
A, The formation and metabolism of vitamin D3. B, Docking studies may explain ligand specificity of cytochrome P450s for vitamin D3 and its metabolites. Top panel, Crystal structure of the Cyp2R1 bound to vitamin D3 (cyan) (PDB ID 3c6g) (100). Middle panel, Homology model of Cyp24A1 (-9.2 kcal/mol) bound to substrate 25(OH)D3 (cyan). Bottom panel, Homology model of Cyp27B1 (-9.4 kcal/mol) bound to 25(OH)D3 (cyan). The heme, heme iron, and bound oxygen (yellow) positions in these cytochrome P450 cavities are shown as spheres at right. The amino acids nearby or in the positions of all three ligands are shown for comparison. Residue names and numbers are provided only if one or more enzyme atoms fall within a 4 Å distance of a ligand atom. The homology models shown were based on the closed ligand cavity conformation observed for the crystal structure of Cyp11A1 (PDB ID 3na0). (All protein structural figures and modeling are courtesy of Dr. James R. Thompson.) C, Physiological changes in response to decreases in serum calcium concentrations.