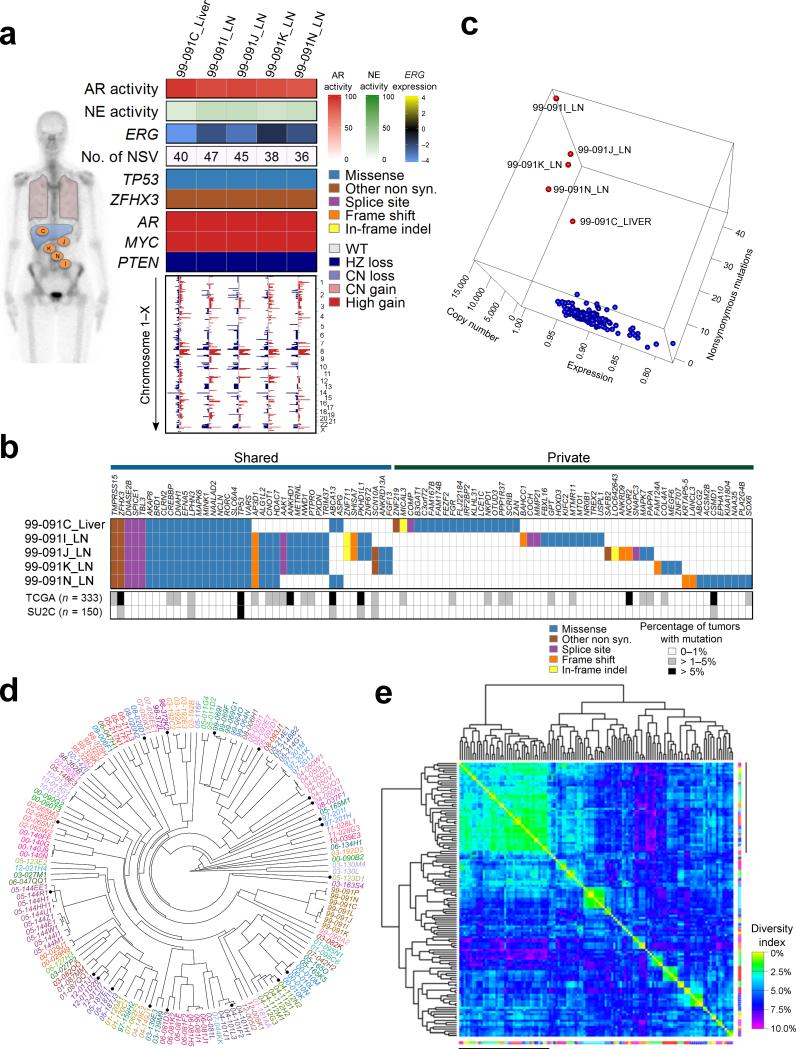
Figure 3. Molecular aberrations are shared between metastasis within individuals with mCRPC.
(a) Gene expression pathways and putative somatic driver aberrations are consistent across metastasis within an individual. Shown are selected features from five metastatic tumors from patient 99-091. Activity scores comprise transcripts of genes regulated by the AR (AR activity score), genes expressed in neuroendocrine carcinoma (NE activity score). ERG transcript levels are relative to the mean-centered ratio for the cohort. NSV is the number of somatic nonsynonymous nucleotide variants. Genome wide copy number losses (blue) and gains (red) are shown for each tumor ordered by chromosome.
(b) Distribution of all 91 nonsynonymous point mutations and indels identified in the five tumors from patient 99-091. The table indicates the presence of a mutation (blue/tan/purple/orange/yellow) or its absence (white) in each tumor. The grey scale bars indicate the frequency of a mutation in the specified gene in the TCGA (primary tumors) or SU2C (metastatic tumors) datasets.
(c) Relationships of tumors derived from a single patient (red points) relative to tumors from all other individuals (blue points) based on a calculated a sample similarity score that comprised single nucleotide alterations, copy number alterations, and gene expression using the 133 tumors with data on all three platforms.
(d) Unsupervised clustering of 149 tumors by correlation of genome-wide copy number variations. Tumors from the same individual are designated with the same color. Primary tumors are noted by a black circle.
(e) Heatmap of the percent of the total of 984 distinct copy number losses and gains identified in the entire cohort of tumors that differ between samples and sorted based on complete linkage dendrograms. Margin line denotes tumors with very low copy number aberration burden. The diversity index is defined as the proportion of corresponding aberrations that differed between samples.