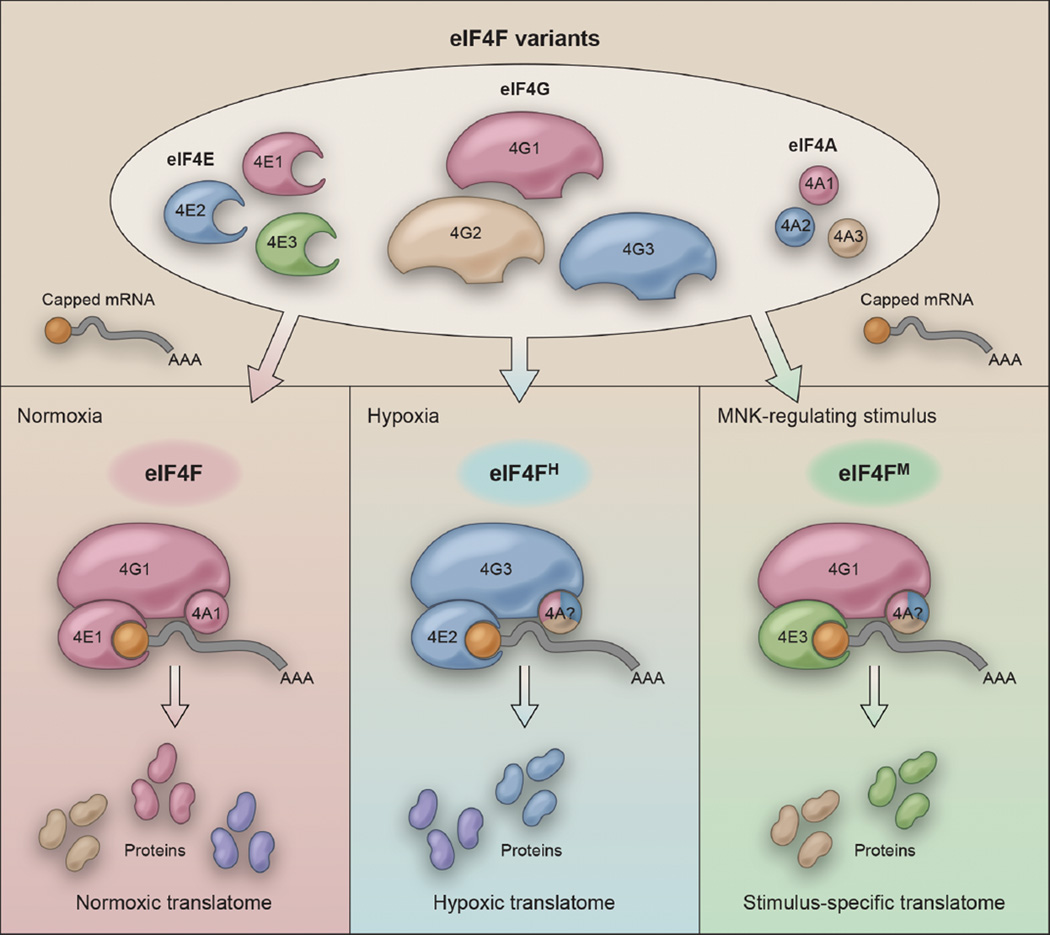
Figure 1. eIF4F variants reprogram the cellular translatome in response to distinct stimuli.
(Top panel) eIF4F-mediated cap-dependent translation in humans involves three major components, eIF4E (cap-binding protein), eIF4G (scaffold protein), and eIF4A (ATP-dependent RNA helicase), each of which has three known homologues (numbered 1, 2, and 3, respectively). Permutations of homologue-specific interactions facilitate adaptation to various stimuli through translatome reprogramming. (Bottom right panel) The basal eIF4F variant contains eIF4G1 and eIF4E1, and produces the normoxic translatome. (Bottom middle panel) Under hypoxic conditions, when eIF4F is inhibited, cells activate the hypoxic eIF4F, eIF4FH, to produce an adaptive, cap-dependent hypoxic translatome. (Bottom right panel) Other stimuli, such as those regulating MNK activity, can potentially activate eIF4E3-containing eIF4F variants (eIF4FM) to produce a stimulus-specific, cap-dependent translatome. The differential utilization of eIF4A variants in distinct eIF4F complexes remains to be tested.