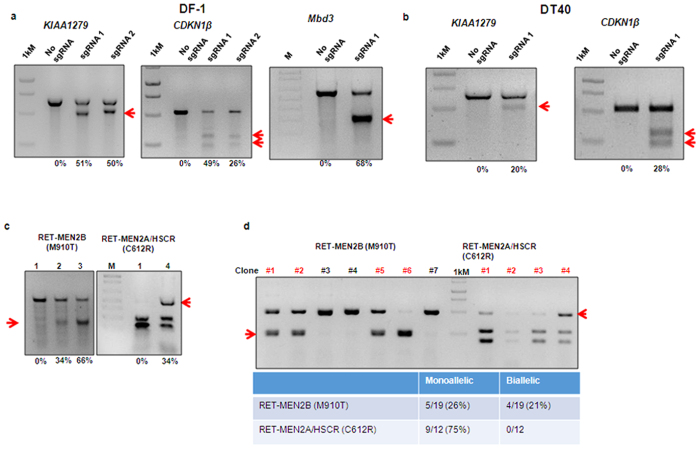
Figure 1. In vitro analysis of NHEJ and HDR genome modification (arrows) mediated by sgRNA-Cas9 system in chicken cell lines.
(a) Frequency (%) of NHEJ mutation mediated by KIAA1279, Cdkn1b and Mbd3-targeting sgRNA-Cas9 system in chicken DF-1 cells by PCR and T7E1 assay. 1kM- 1 kbp DNA ladder, M- 100 bp DNA ladder. (b) Frequency (%) of NHEJ mutation mediated by KIAA1279 and Cdkn1b-targeting sgRNA-CRISPR/Cas9 system in chicken lymphoma B DT40 cells by PCR and T7E1 assay. (c) Representative gel from DF-1 cells transfected with the RET-targeting sgRNA-Cas9 and the ssODN showing efficient integration of the HDR-based BamHI and EcoRV sequence. The frequency of HDR is represented in percentages. 1-No sgRNA, 2- MEN2B sgRNA #1 plus ssODN, 3- MEN2B sgRNA #1 and #2 plus ssODN and 4- MEN2A/HSCR sgRNA 1 plus ssODN. (d) Representative gel for single cell clones derived from DF-1 cells transfected with the RET-targeting sgRNA-Cas9 and the ssODN for the MEN2B and MEN2A/HSCR HDR modifications respectively. The table shows the ratio of the monoallelic and biallelic HDR-based mutations detected with single cell clones and the overall efficiency in percentage: N = 19 for MEN2B and N = 12 for MEN2A/HSCR.