ABSTRACT
Irritable bowel syndrome (IBS) is a chronic functional disorder and its development may be linked, directly and indirectly, to intestinal dysbiosis. Here we investigated the interactions between IBS symptoms and the gut microbiome, including the relation to rifaximin (1200 mg daily; 11.2 g per a treatment). We recruited 72 patients, including 31 with IBS-D (diarrhea), 11 with IBS-C (constipation), and 30 with IBS-M (mixed constipation and diarrhea) and 30 healthy controls (HCs). Of them, 68%, 64%, and 53% patients with IBS-D, IBS-C, and IBS-M, respectively, achieved 10–12 week-term improvement after the rifaximin treatment. Stool samples were collected before and after the treatment, and fecal microbiotic profiles were analyzed by deep sequencing of 16S rRNA, while stool metabolic profiles were studied by hydrogen 1-nuclear magnetic resonance (1H-NMR) and gas chromatography–mass spectrometry (GC-MS). Of 26 identified phyla, only Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria were consistently found in all samples. Bacteroidetes was predominant in fecal samples from HCs and IBS-D and IBS-M subjects, whereas Firmicutes was predominant in samples from IBS-C subjects. Species richness, but not community diversity, differentiated all IBS patients from HCs. Metabolic fingerprinting, using NMR spectra, distinguished HCs from all IBS patients. Thirteen metabolites identified by GC-MS differed HCs and IBS patients. However, neither metagenomics nor metabolomics analyses identified significant differences between patients with and without improvement after treatment.
KEYWORDS: 16s rRNA sequencing, irritable bowel syndrome, metagenomics, metabolomics, rifaximin
Introduction
While infection of the alimentary tract with opportunistic pathogens usually leads to acute gastroenteritis, disruption of the ecological organization of the normal gut microbiota (dysbiosis) may be associated with numerous chronic human disorders, including autoimmune diseases, cancer, inflammatory bowel diseases, obesity, and obesity-linked co-morbidities, such as metabolic syndrome, diabetes, and cardiovascular disorders.1,2
Irritable bowel syndrome (IBS) is a chronic functional disorder, affecting up to 20% of adults in the general population. The diagnosis of IBS is based on a constellation of clinical symptoms, including abdominal pain and/or discomfort, bloating, and distension, accompanied by altered bowel function, ranging from diarrhea-predominant (IBS-D) to constipation-predominant (IBS-C) and the absence of identifiable structural, biochemical, or metabolic abnormalities.3-7 IBS is thought to be due to dysregulation of the brain-gut axis with impaired gut motility and visceral hypersensitivity, impaired gut barrier function and chronic immune activation.8
A treatment with gut-directed antibiotics may profoundly, for a short-term, alter the gut microbiome community structure, but for a long-term usually does not shift the intestine microbiota to a new steady-state. Several studies have investigated a potential role of the intestinal microbiota in the pathophysiology of IBS and found that colonic microbiota may differentiate patients with IBS from healthy controls (HCs).9-16 Treatment with probiotics and antibiotics, including a short course of rifaximin, has improved some IBS symptoms in non-C IBS patients,17-33 providing a direct link between microbiota and IBS. However, some of the IBS patients do not present visible abnormalities in the microbiota composition, and rifaximin may not induce direct changes in the targeted microbiota composition.34 Therefore, alternative mechanisms responsible for rifaximin efficacy, including those directed at the microbiota-gut-brain axis 3,5,7,35,36 should be also considered.
Through the process of fermentation, colonic bacteria produce a wide range of metabolites, which are used as energy sources by epithelial cells in the distal bowel.37-39 These metabolites may also affect the metabolic integrity of intestinal epithelial cells and induce immune responses in the human gut. Different residing gut bacteria can metabolize the same substrates, thereby producing similar metabolites.40-42 The extremely complex and dynamic microbial ecosystem in the gut, especially in the large intestine, may be significantly reduced by its metabolic activity. Thus, the metabolomic testing may be easier than metagenomic testing in determining the clinical end points of dysbiosis. Little is known to date, however, about interactions between the gut microbiome community and metabolites in patients with IBS.40,43
This study analyzed the potential interactions between symptoms attributed to IBS, before and 10–12 weeks after rifaximin treatment, and the gut microbial community by comparing global changes within the microbiotic and metabolic profiles of fecal samples, by sequencing their 16S rRNA and by 1H-NMR and GC-MS techniques.
Results
Clinical analysis and effect size for rifaximin efficacy
We recruited 72 patients, including 31 with IBS-D (diarrhea), 11 with IBS-C (constipation), and 30 with IBS-M (mixed constipation and diarrhea) and 30 healthy controls (HCs). The studied group and the control group did not differ by sex (69% and 66% of females, respectively), age (mean age, 43 and 40 years, respectively) and body mass index (mean BMI (SD), 25.2 (2.8) and 24.4 (4.5), respectively). Overall, 10–12 week-term improvement after rifaximin treatment, defined as similar improvements in symptom severity scores and adequate relief measures for all 4 tested parameters (see Methods), was achieved by 21 (68%), 7 (64%), and 16 (53%) patients with IBS-D, IBS-C, and IBS-M, respectively. The effect sizes in Mann-Whitney paired U-test with power of 80% in a comparison before and after treatment were estimated for 0.53, 0.97 and 0.54 for patients with IBS-D, IBS-C and IBS-M patients, respectively. Therefore, our study had enough power to detect moderate effects for IBS-D and IBS-M, and large effects for IBS-C patients. The effect size in Mann-Whitney paired U-test with power of 80% in a comparison before and after treatment for the whole group of 72 patients was 0.34.
Taxonomy population overview
Amplicons of the 16S hyper-variable regions in bacteria were PCR amplified, and the libraries were sequenced using the PGM platform.44 On average, 90,988 sequences with more than 80% bases of quality of 20 or higher were generated per library. Differentiation of these sequences into operational taxonomic units (OTUs) identified 595 OTUs in Silva taxonomy, as recommended by authors of Mothur.45 Of these, 126 OTUs were identified in more than 0.01% of reads.
OTUs were categorized into subgroups of Phylum, Class, Order, Family, and Genus. Of the 55 known bacterial phyla detected in fecal or mucosal samples from the human gut,46 26 were identified in at least one fecal sample, with only 4 phyla (Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria) consistently found in all samples. The phyla Bacteroidetes and Firmicutes were dominant, with abundances as high as >90% each in different subjects.47 In this study, Bacteroidetes was the predominant phylum in 77%, 69%, and 67% of fecal samples obtained from HCs and from subjects with IBS-D and IBS-M, respectively. By contrast, Firmicutes was the predominant phylum in 91% of samples from IBS-C subjects. Cyanobacteria were detected in more than 80% of samples; Verrucomicrobia were present in more than 50%; and Lentisphaerae, Fusobacteria, Synergistetes, and Tenericutes were observed in more than 20% each. Other phyla were present in fewer than 10% of samples.
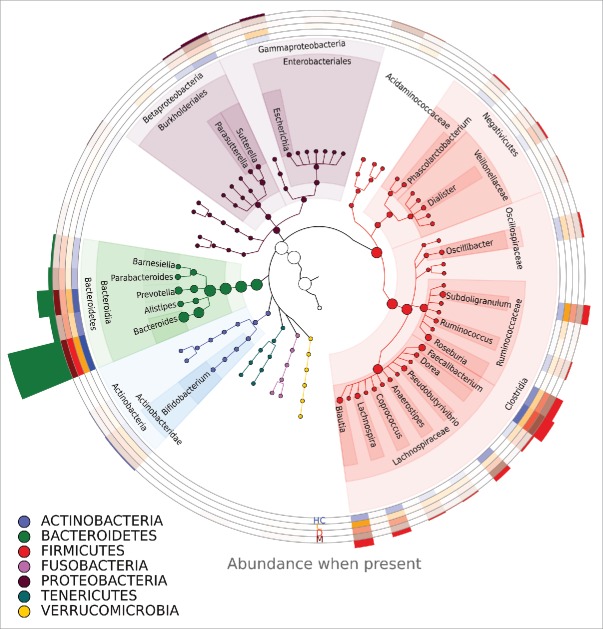
Within the phylum Firmicutes, the class Clostridia was the most prevalent. The two most prevalent families within this class were Lachnospiraceae and Ruminococcaceae. Genera present in all groups within these families included Blautia, Lachnospira, Pseudobutyrovibrio, Roseburia, Subdoligranulum, and Oscilibacter. Within the phylum Bacteroidetes, Bacteroidia was the most prevalent class. The most abundant genera in this order were Bacteroides and Prevotella (Fig. 1).
Figure 1.
The phylogenetic tree of bacteria detected in samples. Only genera present in more than 1% of reads are shown. A more intense color on a heatmap indicates a higher percentage of reads from a given genus. Circles with heatmap represent (from top to bottom) healthy controls, C-, D- and M-type IBS patients before treatment. The histogram above the circles represents the abundance of a genus in all reads. Genera are annotated by their phylum, order, and class or, in the case of the phylum Firmicutes, by their phylum, order, and family.
Taxonomic analysis – IBS subtypes versus healthy controls. Statistically significant differences in Bacteroidetes/Firmicutes ratios were observed between the IBS-C and HC groups and between the IBS-D and IBS-M groups (Fig. 2, Table 1). The most prevalent genera within most samples were Prevotella and Bacteroides, in accordance with enterotypes 1 and 2.48 The Bacteroidetes/Firmicutes ratio in IBS-C patients was lower than in other groups, and the bacteriome of patients with IBS-C was characterized by the prevalence of bacteria from the class Clostridia, especially from families Lachnospiraceae and Ruminococcaceae, in agreement with enterotype 3.48 Principal component analysis (PCA) confirmed differences between healthy individuals and IBS-C patients. By contrast, there were no differences between HCs and the other types of IBS (Fig. S2). Pairwise Mann–Whitney comparisons of HCs and patients with different subtypes of IBS revealed statistically significant differences in 15 taxa between HCs and patients with IBS-C. Statistically significant differences in 2 taxa were observed in HCs and patients with IBS-D.
Figure 2.
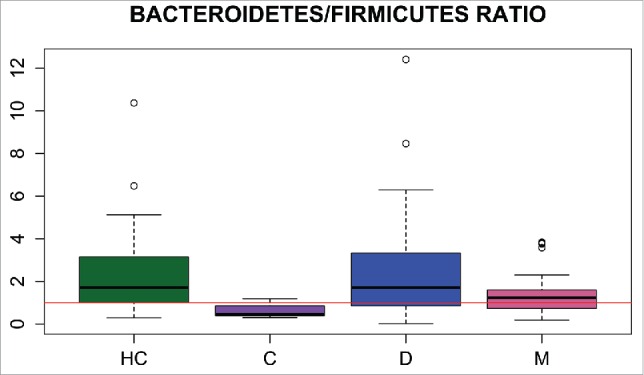
Bacteroidetes/Firmicutes ratios in the healthy control (HC), IBS-C, IBS-D, and IBS-M groups.
Table 1.
Kolmogorov–Smirnov test results for Bacteroidetes/Firmicutes ratio distribution.
| Comparison | q-Value |
|---|---|
| IBS-C/IBS-M | 0.00073 |
| IBS-C/IBS-D | 0.00073 |
| IBS-C/HC | 0.00073 |
| IBS-M/IBS-D | 0.085 |
| IBS-M/HC | 0.162 |
| IBS-D/HC | 0.96 |
Note. HC: healthy control; IBS-D: Diarrhea subgroup; IBS-C: Constipation subgroup; IBS-M: Mixed symptoms subgroup.
Taxonomic analysis – impact of rifaximin treatment. No differences were found in the distribution of Bacteroidetes/Firmicutes ratio before and after the rifaximin treatment (Fig. S1). Seven taxa distinguished patients before and after treatment. However, in any of IBS subgroups there were no taxa discriminating cases before and after treatment.
All the taxa included in these calculations were identified on average in at least 9 reads per sample, suggesting the biological significance of these differences (Table 2).
Table 2.
Mann–Whitney test results showing taxonomic contrasts between healthy controls and IBS-C and IBS-D patients, as well as before and after rifaximin treatment in all IBS patients.
| IBS C-type patients / Healthy controls | |||||
|---|---|---|---|---|---|
| Taxon | Mann-Whitney test statistic | p-Value | Mean abundance- C | Mean abundance - HC | q-Value |
| Bacteroides | 25 | 0.000018 | 0.115 | 0.35 | 0.0022 |
| Coriobacteriaceae | 266 | 0.000310 | 0.00179 | 0.00033 | 0.0155 |
| Ruminococcaceae | 258 | 0.000370 | 0.057 | 0.027 | 0.0155 |
| Clostridiales | 253 | 0.00076 | 0.093 | 0.023 | 0.024 |
| Rhodospirillaceae | 248.5 | 0.00192 | 0.00032 | 0.000035 | 0.038 |
| Clostridiales - Family_XIII_Incertae_Sedis | 249 | 0.00209 | 0.0024 | 0.00059 | 0.038 |
| Granulicatella | 246 | 0.0028 | 0.000178 | 0.000108 | 0.040 |
| Uncultured Ruminococcaceae | 246 | 0.0028 | 0.0199 | 0.0064 | 0.040 |
| Eubacterium | 243 | 0.0031 | 0.000140 | 0.000021 | 0.040 |
| Firmicutes | 240 | 0.0039 | 0.030 | 0.0134 | 0.042 |
| Acetanaerobacterium | 240 | 0.0052 | 0.0028 | 0.00060 | 0.046 |
| Catenibacterium | 237 | 0.0055 | 0.0096 | 0.00035 | 0.046 |
| Lachnospiraceae | 235 | 0.0068 | 0.092 | 0.063 | 0.054 |
| Clostridia | 234 | 0.0076 | 0.0035 | 0.00144 | 0.056 |
| Parabacteroides | 65 | 0.0083 | 0.0108 | 0.025 | 0.058 |
| IBS D-type patients / Healthy controls | |||||
| Taxon |
Mann-Whitney test statistic |
p-Value |
Mean abundance - D |
Mean abundance - HC |
q-Value |
| Porphyromonadaceae | 244 | 0.00070 | 0.00158 | 0.0040 | 0.053 |
| Alistipes | 248 | 0.00087 | 0.026 | 0.049 | 0.053 |
| IBS patients before and after treatment (paired test) | |||||
| Taxon |
Mann-Whitney test statistic |
p-Value |
Mean abundance – before treatment |
Mean abundance – after treatment |
q-Value |
| Bilophila | 564 | 0.000201 | 0.0028 | 0.0043 | 0.025 |
| Clostridiales | 1935 | 0.00050 | 0.030 | 0.021 | 0.027 |
| Catabacter | 1328 | 0.00084 | 0.00030 | 0.000193 | 0.027 |
| Parasutterella | 621 | 0.0032 | 0.0059 | 0.0097 | 0.055 |
| Clostridiales - Family_XIII_Incertae_Sedis | 1745 | 0.0033 | 0.00070 | 0.00034 | 0.055 |
| Clostridiales - Family_XIII_Incertae_Sedis uncultured | 1790 | 0.0034 | 0.00048 | 0.00029 | 0.055 |
| Firmicutes | 1834 | 0.0036 | 0.0155 | 0.0106 | 0.055 |
Diversity analysis
Species richness (i.e., the total number of species per sample), estimated by Chao1, differed between all patients with IBS and HCs, as well as differing before and after treatment (Fig. 3). However, community diversity (i.e., the evenness of species distribution), as estimated by the Simpson index, did not differ between the studied groups (Fig. 4).
Figure 3.
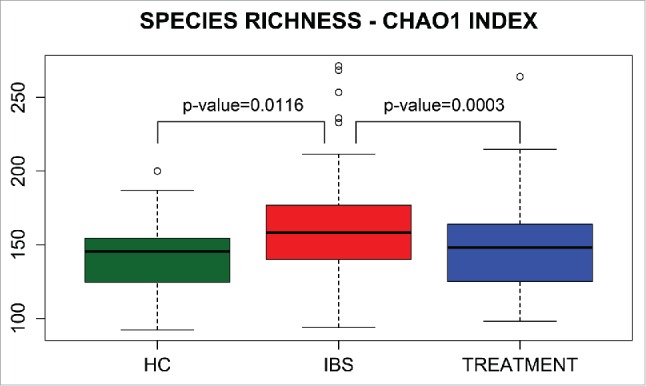
Boxplots of Chao1 species richness index in healthy controls (HCs) and in IBS patients before (IBS) and after (Treatment) treatment.
Figure 4.
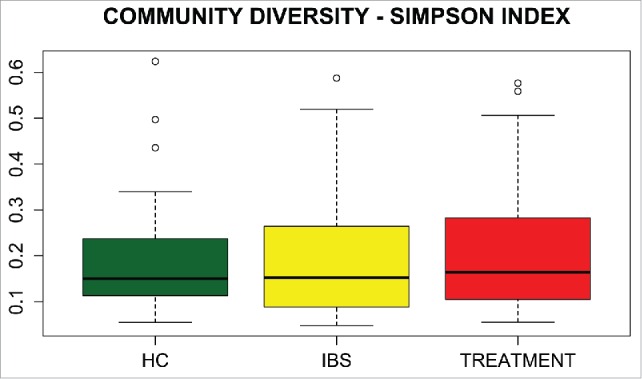
Box plots of the Simpson index of community diversity in healthy controls (HCs) and IBS patients before (IBS) and after (Treatment) treatment.
Functional analysis
While microbial abundances within the same habitat varied widely among subjects, the distribution of pathways representing processes for microbial life was much more consistent.47 To gain more insight into metabolic functions related to bacterial activity, the bacterial taxa were assigned to KEGG metabolic pathways using the Greengenes reference dataset. The IBS-C group differed markedly from both the other groups of IBS patients and HCs (Fig. S3). Those differences include those involved in the most abundant metabolic pathways, including alanine, aspartate, and glutamate metabolism; amino sugar and nucleotide sugar metabolism; and oxidative phosphorylation, all of which are under-represented in the metagenome of IBS-C patients. By contrast, methane metabolism and pyruvate metabolism were over-represented in IBS-C patients (Fig. S4). No significant differences were observed in the presence and abundance of metabolic functions between IBS patients before and after rifaximin treatment and between patients who did and did not exhibit short-term improvement after treatment.
Metabolic fingerprinting of fecal samples from IBS patients
Representative 1H-NMR spectra of chloroform extract of feces of HCs and IBS patients are shown in Figure S5. Because the number of patients was relatively small, a cross-validated 2-group partial least-squares-discriminant analysis (PLS-DA) was performed to determine any possible between-group differences. The obtained parameters of analysis from all studied comparisons are collected in Table S1. Of these, only the comparison of HCs and all IBS patients before rifaximin treatment was significant (Table S1), with visible differences observed on the PLS-DA plots (Fig. 5). Despite the very low parameters of the remaining models, distinct trends separating IBS subgroups, especially IBS-C, and HCs may be present (Table S1, Fig. 6). However, this analysis could not differentiate between patients who did and did not improve after rifaximin treatment.
Figure 5.
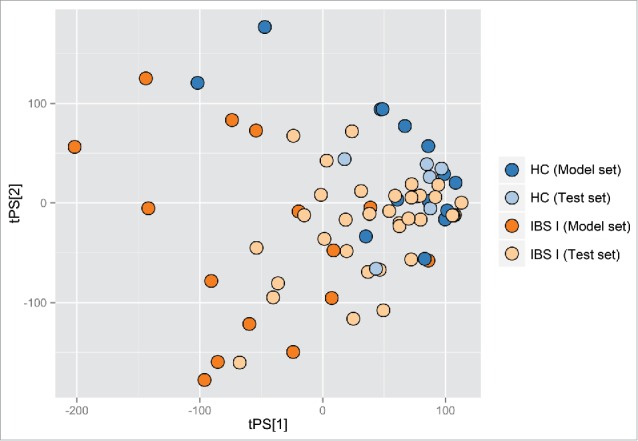
Partial least-squares-discriminant analysis score plot based on metabolic fingerprints for chloroform extracts of stool samples. HC- healthy control; IBS I - IBS patients.
Figure 6.
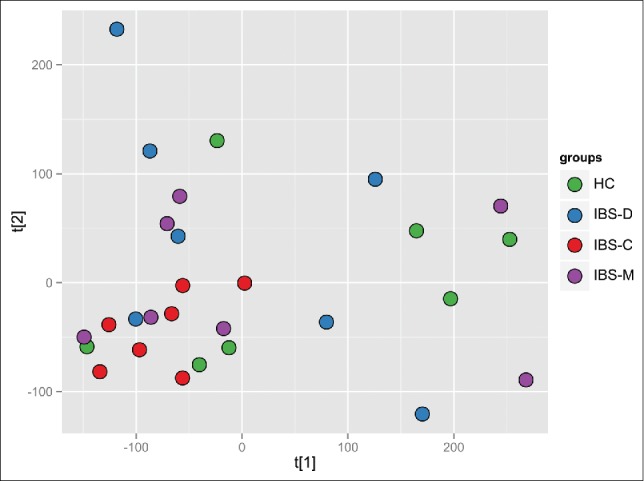
The PCA plot based on the metabolic fingerprints for chloroform extracts of stool samples, comparison between healthy control (HC) – green and IBS patients before treatment divided into subgroups.
GC-MS analysis of smaller numbers of samples (8 HCs and 29 IBS patients) allowed the identification of 1174 metabolites. Of these, 13 exhibited statistically significant (false discovery rate - FDR, ≤0 .1, and FC, ≥1 .5) differences in abundance in a simultaneous multi-group comparison of HCs and the 3 subgroups of IBS patients before treatment (Table S2). In addition, 13 metabolites differed significantly in HC samples and in samples from the 3 subgroups of IBS patients after treatment (Table S3). Of these, 8 metabolites were common for the 2 comparisons. As shown on the PCA plots (Figures S6 and S7), the differences were due to differences in abundance between HCs and the IBS-M and IBS-D subgroups, while the dissimilarity of IBS-C samples was less evident.
These results suggest that rifaximin treatment has a limited effect on metabolite contents of stool. Although changes in the abundance of 2 compounds (Propanoic acid, 2-(methoxyimino)-, trimethylsilyl ester and Nonanoic acid, trimethylsilyl ester) were statistically significant (unadjusted p-values ≤0.05) in a paired-sample comparison of patients before and after treatment, they failed to meet the established criterion of FDR. No significant differences were observed between patients with and without improvement after treatment.
Discussion
The human microbiome consists of a variety of bacteria, archaea, fungi, and viruses.47 By 12–18 months of age, an infant's intestine is colonized by more than 1,000 species, normally commensal or mutualists. The relatively stable composition of gut microbiota within individuals is modulated by many factors, including diet, sanitation, antibiotics, and aging.49 This complex ecosystem trains the immune system, protects against opportunistic pathogens, harvests nutrients and energy from the diet, and ferments non-digestible carbohydrates.7 Understanding the interactions among microbiome-associated diseases and dysbiosis may enable prevention and treatment, by restoring a healthy microbial community in a personalized way.
While the diagnosis of IBS is largely subjective and based on symptoms, consisting mostly of abdominal pain and changes in bowel habits, functional bowel symptoms are common in the general population and vary over time.50 Due to the great symptom variability among individuals and subgroups of patients with IBS, identification of specific microbial groups whose relative abundance can contribute to the disease and respond to treatment is challenging.
Research of complex microbial ecosystems requires adequate methods to document bacterial presence/absence and abundance. Analytical revision of studies assessing the association between intestinal microbial profiling and IBS 14 has shown that culture-based techniques allow identification of only a small proportion of gut colonizers. By contrast, bacterial identification based on the taxonomically informative 16S rRNA gene sequences (culture-independent techniques) provide a more global picture of gut microbial configuration but, despite this, no standardized procedure for metataxonomic approach has been accepted so far. While some investigators prefer to sequence amplicons of individual hypervariable regions of the 16S rRNA gene, including V1, V3, V4 or V5, others employ sequencing of a single amplicon spanning 2 or more regions, eg. V1-V2,51 V1-V3.52 In a consequence, an arbitrary choice of the region(s) for a library creation may lead to an amplification bias which results from, among other causes, an insufficient specificity of primer annealing to capture relevant bacterial taxa.52 This study utilized a sequencing protocol for the PGM platform using an Ion 16S Metagenomics Kit that allows a consensus view across 6 regions (V2, V3, V4, V6-7, V8 and V9) and, as we showed recently, it reliably captures and quantifies the composition of a reference mock community.44
Altogether, 595 OTUs were identified in fecal samples using Silva taxonomy. Of 26 identified phyla, only 4, Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria, appeared to be universal, being present in all samples. The Bacteroidetes/Firmicutes ratio in IBS-C patients was lower (Fig. 2) than that in other groups. In agreement with previous results,53 this study showed that species richness was significantly higher in the whole group of IBS patients than in controls (Fig. 3), although community diversity did not differ (Fig. 4). Lack of differences in community diversity, previously reported for example by Krogius-Kurikka et al.54 or Carroll et al.55 studies, may stem from differences in 16S rRNA sequencing approach: most methods focus on one or 2 hypervariable regions, while kit we used covers 6 regions.44 Pairwise Mann–Whitney testing comparing healthy controls and patients with different subtypes of IBS revealed statistically significant differences in 18 taxa between HCs and the IBS-C subgroup and in 2 taxa between HCs and the IBS-D subgroup, respectively (Table 2). The gut bacteriome of patients with IBS-C was characterized by the prevalence of bacteria from the class Clostridia (Fig. 1).
Depending on the medium used in culture-based protocols, either reductions in Bifidobacteria56,57 or no differences in their concentration15,58,59 were observed in stool samples from IBS patients, while the level of the genus Lactobacillusis was either increased60 or reduced57,59 in IBS fecal samples. However, since 80% to 99% of the microorganisms from any environment are not cultivable,61 the significance of these data is reduced.
A clone library-based method, analyzing the sequence of a limited number of 16S rRNA gene clones, found that the prevalence of Clostridium spp. was increased and the prevalence of Eubacterium was decreased in IBS patients.15 By contrast, another study showed no significant differences between the microbiota compositions of both duodenal biopsies and fecal samples from IBS patients and HCs, except for an increase of Pseudomonas aeruginosa in IBS.62 Research using differential centrifugation to separate genomic DNA from fecal samples 16 detected reduced members of Lactobacillus in all IBS subgroups, less abundant Actinobacteria in IBS-C and IBS-D patients, higher levels of Ruminococcus in IBS-C and IBS-M patients, and higher levels of Streptococcus in IBS-D patients. The sequences of a much larger number of clones (3267) revealed increases in Proteobacteria and Firmicutes (especially the family Lachnospiraceae), and decreases in Actinobacteria and Bacteroidetes in IBS-D patients, with decreased bacterial diversity.54
Finally, the high-throughput pyrosequencing of the variable regions V1–V3 (an average of 8232 reads per sample) and V6 (an average of 6591 reads per sample) of the 16S rRNA gene showed less microbial richness, greater abundance of the phylum Proteobacteria, and lower abundance of the genus Faecalibacterium and the species Faecalibacterium prausnitzii in IBS-D patients.55 Pyrosequencing also showed that the abundances of γ-Proteobacteria (particularly, the species Haemophilus parainfluenzae) and of the Firmicutes genera Dorea and Veillonella were increased in pediatric IBS patients.63 The genus Veillonella was also increased in pediatric IBS-D patients.64 An analysis of about 268,000 reads from 16S rRNA genes by pyrosequencing65 showed reduced microbial diversity in IBS samples, high abundances of Rikenellaceae, Porphyromonadaceae, and Bacteroidaceae, and reductions in Ruminococcaceae spp.
In conclusion, the obtained data appeared rather weakly consistent, partly as a result of different protocols employed to investigate gut ecosystems. Both the quality and significance of the results may be influenced by the inter-individual variability in the gut microbiome, which could be attributed to different IBS symptoms. The limitations of this study were due primarily to the HC group, which consisted mostly of hospital employees. This group may not accurately represent the bowel habits, daily activities, and stress levels of the general population.50 However, the selection of the control group in the study on intestinal microbiome in IBS patients is particularly challenging keeping in mind that etiology of functional disorders is mainly unknown and their diagnosis is based on clinical findings, on one hand, while diversity of gut microbiota is modified by a plurality of factors, on the other hand. Despite this, our taxonomic findings in IBS patients are in good agreement with work by Soldi et al.,34 who in 15 non-constipated IBS subjects, treated with rifaximin at daily dose of 1650 mg for 14 days, have observed effective relief of IBS symptom without changes of the overall composition of the core microbiota, even at the end of treatment, although they have found some fluctuations in a few bacterial groups.
Symptoms attributed to IBS may be more frequent after an episode of gastroenteritis and may be caused by small intestinal bacterial overgrowth (SIBO).66 Multiple controlled trials have confirmed the effectiveness of both systemic antibiotics and non-systemic rifaximin (administrated at daily dose ranging between 800 and 1650 mg for 10 – 14 days) in SIBO eradication as well as an improvement of IBS global symptoms 30-32,67-72 that persisted ≥12 weeks post treatment. In addition to clinical studies, oral rifaximin in rats altered the composition of bacterial communities in the ileum and prevented mucosal inflammation, impairment of intestinal barrier function, and visceral hyperalgesia in response to chronic stress.37 In accordance to the American College of Gastroenterology recommendations, rifaximin was approved in 2015 for the treatment of IBS with diarrhea.28
Of our patients with IBS-D, IBS-C, and IBS-M who received rifaximin (1200 mg/day for approximately 10 days), 66%, 64%, and 53%, respectively, experienced improvements in IBS symptoms 10–12 weeks after treatment. Thus, the symptom improvement was achieved not only in IBS-D, but also in IBS-C patients. However, although rifaximin treatment significantly lowered the increased species richness in fecal samples from IBS subjects (Fig. 3), and 8 taxa distinguished the entire group of patients before and after the rifaximin treatment (Table 2), no differences in OTU abundance were observed between IBS patients who did and did not experience short-term improvement after treatment. The rifaximin appeared to be efficacious without inducing dramatic changes in gut microbiome not only in IBS patients (34 and this study), but also in patients with hepatic encephalopathy.73,74
As the species richness suggests (Fig. 3), rifaximin may act on low-abundance organisms that contribute only marginally to the overall gut community but can be associated with various symptoms of IBS. Because the microbial variation between individuals is greater than that of samples from the same subject at different points in time, these low-abundance colonizers may be overshadowed by dominant ones, especially when assessing a relatively small number of patients, resulting in underpowering of microbial presence and abundance. Additional studies of microbial groups whose abundance is related to the variability among IBS symptoms and response to treatment may uncover roles for these low-abundance taxa.
Cooperation between the gut microbiome and mammalian metabolism is an essential element of normal gastrointestinal function. The gut bacteria are able to break down indigestible food components and produce essential metabolites, including short chain fatty acids (SCFAs), branched chain fatty acids (BCFAs), amino acids, carbohydrates (predominantly glucose), phenolics, (poly)amines, bile acids, and glycerol. Although the amounts of these products are quite variable in fecal extracts, they may characterize gut dysbiosis and its related metabolic activities.38,42,75 Thus, metabolomic studies may help understand the ethiopathological mechanisms of gastrointestinal alterations and uncover the diagnostic value of related metabolomic biomarkers.
Previous studies showed an increase in abundance of the cyclic ester 2(3H)-furanone and slightly reduced levels of dodecanoic, azelaic, and adipic acids in the mucosa of IBS patients,41 while the fecal metabolic profile of patients with IBS revealed increased butyrate and reduced acetate and propionate,76 or increased acetate and propionate, with unchanged butyrate.77 Changes in fecal esters of SCFAs, cyclohexanecarboxylic acid and its ester derivatives were associated with IBS-D,42 and significant reductions of BCFAs were observed.38 The unbalanced fecal organic acid levels in IBS correlated with the altered profile of intestinal microbiota, especially Lactobacilli and Veillonella.60
1H-NMR spectroscopy represents a powerful technique for investigating gut metabolomic profiling, with the simplicity of sample preparation and the high throughput being its major benefits.75 Our fingerprint profiling of the lipophilic metabolites in fecal samples from IBS patients provided a 2-group PLS-DA model distinguishing IBS patients from HCs (Fig. 5). However, the relatively small number of patients in each IBS subgroup did not permit the differentiation among all 4 studied groups (IBS-D, IBS-M, IBS-C, and HCs) using one model, or between each IBS subgroup and HCs using the 2-group PLS-DA model. Also, fingerprint profiling did not differ significantly in IBS patients before and after rifaximin treatment. Nevertheless, trends were detected toward distinguishing IBS-C from the 2 other IBS subgroups and controls (Fig. 6). The direct causal link between the microbial composition and the corresponding fecal metabolite profiles suggests that the low discrimination potential of metabolomic profiles in IBS patients confirms the rather subtle dysbiosis in IBS patients.
GC-MS showed that, of 1174 identified metabolites, 13 exhibited statistically significant differences in abundance in fecal samples from HCs and IBS patients before treatment (Table S2), with another 13 differing significantly after treatment (Table S3). However, the effects of rifaximin on metabolite levels were limited, with no significant differences between patients who did and did not show improvement after treatment.
Although definitive microbiological signatures of IBS have not been established, previous metagenomic studies consistently showed that IBS is associated with gut dysbiosis, and that antibiotics and probiotics may be beneficial in treatment. Probiotics and antibiotics targeting colonic microbiota improved some IBS symptoms, suggesting a direct link between microbiota and IBS. Alterations in intestinal microbiota may also indirectly link the development and maintenance of IBS with impairment of the microbiota-gut-brain axis. Thus, dysbiosis may not only be a consequence of IBS, but a plausible causative factor.14 However, the relationship between human gut microbiota and IBS is still not well understood, and further experimental research is required. Although symptoms attributed to IBS may be more frequent after an episode of gastroenteritis66 and may be caused by SIBO, no difference among major phyla or genera were found in small intestine microbiota.78
Our study showed rather discrete IBS-related alterations of both the fecal microbiome and metabolome. The observed differences among the taxonomy of gut microbiota in IBS patients may reflect difficulties in classification of IBS subtypes and variability in IBS patient cohorts, but may also be due to differences in methodology and the lack of statistical power of the research. Thus, the relationships of IBS with gut microbiota composition and metabolite production are still undetermined, and it is unclear whether IBS is a disorder of the small intestine, large intestine, or both.78 Although analyses of dysregulation of colonic microbiota and their metabolic activities cannot be currently employed in clinical practice, further studies may identify candidate bacteria and/or metabolites that are practically useful.
Methods
Patients
From August 2012 to February 2014, 72 IBS patients (50 females and 22 males), the average age(SD): 43(13) and 30 HCs (20 females and 10 males), the average age (SD): 40(12) were recruited into this study by 2 gastroenterologists with expertise in IBS (RT and JO). All patients met symptom-based Rome III diagnostic criteria for IBS, and Bristol Stool Form Scale was used to identify alteration types in the patients' gut transit.79 Accordingly, patients were classified as suffering from IBS-D, IBS-C or IBS-M. Control individuals, mostly hospital employees, reported themselves as “healthy.” The IBS patients were among those admitted to the Gastroenterological Outpatient Departments for severe abdominal bloating and fullness that considered the main indications of choice for treatment with 1200 mg/day rifaximin for approximately 10 d (the total dose was 11.2 g per a treatment). Patients were asked not to use other treatment until the second stool sample was collected, including laxatives, pre- and probiotics. IBS patients with a history of inflammatory bowel disease (IBD), severe cardiovascular and/or respiratory disease, and/or renal disease, as well as those being treated with antibiotics, corticosteroids, or IBS prescription medications, were excluded from the study.
During medical interviews at the initial visit and 10–12 weeks after the treatment, patients were asked to fill out questionnaires regarding the following: 1) their bowel function and habits; 2) their degree of recurrent abdominal pain, rated on a 5-point scale, from discomfort without pain to very severe pain; 3) the degree of abdominal bloating and fullness, rated on a 4-point scale, from not at all to extremely; and 4) the impact of IBS symptoms on quality of life, rated on a 4-point scale from not at all to significant deterioration. In addition, symptom improvement was also assessed using a dichotomous measure with a single question: “Did you have adequate relief of the relevant symptom?”. 14 The improvement was defined when similar improvements in symptom severity scores and adequate relief measures for all 4 tested parameters was reported by the patient.
All participants were unrelated Polish Caucasians who lived in the urban Mazovia region of Poland, mainly in the Warsaw agglomeration. The study was approved by the local ethics committee (Cancer Center-Institute, Warsaw, Poland), and informed written consent was obtained from all subjects. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki.
Stool collection and preparation
Subjects were provided with a stool specimen collection kit, consisting of a styrofoam box, tubes, and spatulas for stool samples, and an ice pack, and a disposable bag at the initial screening visit. Stool samples from a single bowel movement were collected by each subject before and 10–12 weeks after treatment with rifaximin. Collected stool samples were immediately frozen in a home freezer and kept at −20°C. Aseptic techniques using a disposable scalpel were utilized to scrape off approximately 200 mg of each stool sample for DNA purification, and 100 mg of stool were added to 2 2 mL Eppendorf tubes for metabolome analysis.
DNA extraction
DNA was isolated from stool samples using QIAamp DNA Stool Mini Kits (Qiagen). Briefly, 1 mL of InhibitEX Buffer was added to an Eppendorf tube containing 200 mg of the stool sample. The tube was vortexed thoroughly until the suspension was homogenized. The sample was heated at 95°C for 5 min and centrifuged. A 200 µL aliquot of supernatant was transferred to a fresh tube, mixed with 15 µL of Proteinase K and 200 µL of AL buffer, and incubated at 70°C for 10 min. Ethanol (200 μL) was added to each tube, and DNA was recovered on QIAamp spin columns according to the QIAamp DNA Stool Kit protocol. DNA samples were eluted and stored in Tris-HCl buffer, pH 8.0, at −20°C.
16S rRNA sequencing
DNA was sequenced on a PGM platform using Ion 16S Metagenomics Kit (Life Technologies; A26216) as described before.44
Identification of bacterial taxa
Unmapped bam files from the PGM were converted into fastq with SamToFastq script (Picard Tools version 1.115 ),80 and the sequences were filtered with a fastq_quality_filter from FASTX-Toolkit (version 0.0.13),81 so that only sequence with more than 80% bases of quality 20 (on the Phred scale) or higher remained. Further steps of the analysis were performed with Mothur (version 1.34.0).45 Fastq files were converted into fasta format. The 16S rRNA sequences were classified by the Wang method, using the Silva bacterial 16S rRNA database as a template (release 102, retrieved from Mothur wiki page) and 60% as the value for bootstrap cut-off. Bacteria were classified according to Silva taxonomy, and taxonomic profiles were created with modified script from STAMP (version 2.0.8).82
Data visualization and statistical analysis
Data visualization, including percentages of bacterial taxa in each sample, statistical tests, and the PCA, was performed in R (version 3.1.1) and graphics package ggplot2 (version 1.0.1).83 Differences among Bacteroidetes/Firmicutes distribution ratios were compared using the Kolmogorov–Smirnov test. Taxonomic differences among groups were determined using Mann–Whitney U-tests, whereas differences before and after antibiotic therapy were analyzed by Mann–Whitney paired tests. Taxa with essentially constant abundance (log2 (IQR) <0.5) were removed from taxonomic analyses. P-values were corrected for multiple hypothesis testing using the Benjamini–Hochberg procedure 84 to control the FDR. The power analysis was conducted in GPower 3.1.85
Biological diversity analysis
The species richness Chao1 index and community diversity Simpson index were computed in Mothur. Differences between groups were determined using Student's t-tests, whereas differences before and after antibiotic therapy were analyzed by Student's paired t-tests.
Metabolic pathway analysis
Taxa were assigned to Greengenes taxonomy, using mothur, and then to KEGG Pathways with PICRUSt (version 1.0.0).86 For further analysis, only metabolic pathways were considered. Functional differences between groups were calculated using Mann–Whitney U-tests, whereas differences before and after antibiotic therapy were calculated using Mann–Whitney paired tests. P-values were corrected for multiple hypothesis testing using the Benjamini–Hochberg procedure to control the FDR.
Extraction of metabolites
One 100 mg stool sample was added to an Eppendorf tube containing 1 mL of methanol, and another 100 mg stool sample was added to a tube containing chloroform. The samples were vortexed at 1400 rpm for 1 h in a ThermoMixer (Eppendorf) at room temperature, followed by centrifugation for 10 min at 10,000 rpm at room temperature. The supernatants were decanted and again centrifuged, and the final supernatants were transferred to fresh tubes and evaporated to dryness in a CentriVap centrifugal vacuum concentrator.
NMR analysis
Pellets from chloroform extracts (see above) were immersed in 600 µL of deutered chloroform, and 550 μL of each sample was transferred to a 5 mm NMR tube. All NMR spectra were recorded at 300 K using a Bruker Avance II 600 spectrometer (Bruker GmBH, Germany) operating at a proton frequency of 600.58 MHz with the following parameters: relaxation delay, 3.5 s; acquisition time, 2.48 s; number of transients (scans), 40; number of points, 64 K; pulse program, zgpr1d (in Bruker notation) with chloroform presaturation; spectral width, 20 ppm; and line-broadening factor, 0.3 Hz. The spectra were manually corrected for phase and baseline distortions and were referenced to the tetramethylsilane (TMS) signal (δ = 0.00 ppm).
Statistical analysis of NMR results
Data were processed, and multivariate statistical data were analyzed as described.87 All spectra were exported to Matlab (Matlab v. 8.1, MathWorks, Inc.). Regions affected by solvent suppression were excluded, and signal alignment procedures involving correlation optimized warping (COW) and interval correlation shifting (icoshift) algorithms were applied. Fecal spectra each consisted of 10,000 data points, which were normalized using the Probabilistic Quotient method (PQM) to overcome the issue of dilution. Prior to chemometric analysis, the data set was Pareto (Par) scaled. The differences in metabolite fingerprints were assessed using preliminarily PCA and then least-squares-discriminant analysis (PLS-DA) with representative samples selection by use of Kennard-Stone algorithm for both types of analysis.88 A default 7-fold cross validation (CV-ANOVA) was applied to each PLS-DA model (one/seven of the samples being excluded from calculations in each round).
GC-MS analysis
Methanol extracts were evaporated using a vacuum concentrator (Labconco), and the samples were further dried under vacuum and over P2O5, and derivatized to block polar groups of compounds present in the mixture. Compounds were derivatized by incubation for 1.5 h at 37°C with 100 µL of 20 mg/mL methoxyamine hydrochloride in pyridine, followed by incubation with 160 µL of N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA) for 30 min at 37°C.
Samples were qualitatively and quantitatively assayed using a LECO Pegasus 4D system, consisting of a 7890A gas chromatograph (Agilent) and a LECO ToF mass analyzer. Data were analyzed using LECO ChromaTOF software version 4.51.6.0. Gas chromatography was performed using a 30 m long, 0.25 mm internal diameter DB-5MS column with 0.25 µm film thickness (J&W Scientific, Agilent). For injection, a Gerstel CIS PTV-type injector was used. The injection temperature was 40°C, increasing 10°C/sec to 240°C, with the MS transfer line and ion source set at 250°C. Pure helium was used as the carrier gas at a constant flow of 1 mL/min. The oven temperature was held constant at 70°C for 2 min, increased 10°C/min to 300°C, and held constant for 10 min at 300°C. Mass spectra were recorded in a range of 35–650 m/z in EI + mode under standard 70 eV ionization conditions. The retention index mixture was run prior to relevant analyses, and an appropriate Retention Index Method was created based on that. Peaks were identified based on their retention indices and comparisons of their spectra with those in proper mass spectra databases (NIST).
Statistical analysis of GC-MS results
Normalized peak areas of metabolites were log-transformed and imported into MStat, a statistical analysis software tool running in the Matlab environment (available at http://proteom.ibb.waw.pl/mstat). For multiple group comparisons, an ANOVA-based resampling significance test was used. Paired comparisons were performed using a resampling test with paired-sample t statistics. In both cases, the resulting p-values were corrected for multiple hypothesis testing using the Benjamini–Hochberg procedure to control the FDR. Only abundance changes with FDR-adjusted p-values ≤0.1 and fold-change (FC) values ≥1.5 were considered significant. PCA was used to graphically evaluate the relationships among the studied samples.
Supplementary Material
Abbreviations
- IBS
irritable bowel syndrome
- SIBO
small intestinal bacterial overgrowth
- 1H-NMR
1-nuclear magnetic resonance
- GC-MS
gas chromatography–mass spectrometry
- HC
healthy control
- OTU
operational taxonomic unit
- PCA
principal component analysis
- FDR
false discovery rate
- BMI
body mass index
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Accession codes
Deep sequencing data have been deposited at The European Bioinformatics Institute (EBI) Metagenomics repository under accession number PRJEB11252.
Funding
This study was funded by the National Science Center, grant number 2011/03/B/NZ5/01511.
References
- [1].Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology 2009; 136:65-80; PMID:19026645; http://dx.doi.org/ 10.1053/j.gastro.2008.10.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fujimura KE, Slusher NA, Cabana MD, Lynch SV. Role of the gut microbiota in defining human health. Expert Rev Anti Infect Ther 2010; 8:435-54; PMID:20377338; http://dx.doi.org/ 10.1586/eri.10.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hyland NP, Quigley EM, Brint E. Microbiota-host interactions in irritable bowel syndrome: epithelial barrier, immune regulation and brain-gut interactions. World J Gastroenterol 2014; 20:8859-66; PMID:25083059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cuomo R, Andreozzi P, Zito FP, Passananti V, De Carlo G, Sarnelli G. Irritable bowel syndrome and food interaction. World J Gastroenterol 2014; 20:8837-45; PMID:25083057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ford AC, Quigley EM, Lacy BE, Lembo AJ, Saito YA, Schiller LR, Soffer EE, Spiegel BM, Moayyedi P. Efficacy of prebiotics, probiotics, and synbiotics in irritable bowel syndrome and chronic idiopathic constipation: systematic review and meta-analysis. Am J Gastroenterol 2014; 109:1547-61, 1562; http://dx.doi.org/ 10.1038/ajg.2014.202 [DOI] [PubMed] [Google Scholar]
- [6].Drossman DA. The functional gastrointestinal disorders and the Rome III process. Gastroenterology 2006; 130:1377-90; PMID:16678553; http://dx.doi.org/ 10.1053/j.gastro.2006.03.008 [DOI] [PubMed] [Google Scholar]
- [7].Lee KN, Lee OY. Intestinal microbiota in pathophysiology and management of irritable bowel syndrome. World J Gastroenterol 2014; 20:8886-97; PMID:25083061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Drossman DA, Camilleri M, Mayer EA, Whitehead WE. AGA technical review on irritable bowel syndrome. Gastroenterology 2002; 123:2108-31; PMID:12454866; http://dx.doi.org/ 10.1053/gast.2002.37095 [DOI] [PubMed] [Google Scholar]
- [9].Bradley HK, Wyatt GM, Bayliss CE, Hunter JO. Instability in the faecal flora of a patient suffering from food-related irritable bowel syndrome. J Med Microbiol 1987; 23:29-32; PMID:3820268; http://dx.doi.org/ 10.1099/00222615-23-1-29 [DOI] [PubMed] [Google Scholar]
- [10].Si JM, Yu YC, Fan YJ, Chen SJ. Intestinal microecology and quality of life in irritable bowel syndrome patients. World J Gastroenterol 2004; 10:1802-5; PMID:15188510; http://dx.doi.org/ 10.3748/wjg.v10.i12.1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Malinen E, Rinttilä T, Kajander K, Mättö J, Kassinen A, Krogius L, Saarela M, Korpela R, Palva A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol 2005; 100:373-82; PMID:15667495; http://dx.doi.org/ 10.1111/j.1572-0241.2005.40312.x [DOI] [PubMed] [Google Scholar]
- [12].Lyra A, Rinttilä T, Nikkilä J, Krogius-Kurikka L, Kajander K, Malinen E, Mättö J, Mäkelä L, Palva A. Diarrhoea-predominant irritable bowel syndrome distinguishable by 16S rRNA gene phylotype quantification. World J Gastroenterol 2009; 15:5936-45; PMID:20014457; http://dx.doi.org/ 10.3748/wjg.15.5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Codling C, O'Mahony L, Shanahan F, Quigley EM, Marchesi JR. A molecular analysis of fecal and mucosal bacterial communities in irritable bowel syndrome. Dig Dis Sci 2010; 55:392-7; PMID:19693670; http://dx.doi.org/ 10.1007/s10620-009-0934-x [DOI] [PubMed] [Google Scholar]
- [14].Taverniti V, Guglielmetti S. Methodological issues in the study of intestinal microbiota in irritable bowel syndrome. World J Gastroenterol 2014; 20:8821-36; PMID:25083056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mättö J, Maunuksela L, Kajander K, Palva A, Korpela R, Kassinen A, Saarela M. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome–a longitudinal study in IBS and control subjects. FEMS Immunol Med Microbiol 2005; 43:213-22; http://dx.doi.org/ 10.1016/j.femsim.2004.08.009 [DOI] [PubMed] [Google Scholar]
- [16].Kassinen A, Krogius-Kurikka L, Mäkivuokko H, Rinttilä T, Paulin L, Corander J, Malinen E, Apajalahti J, Palva A. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterol 2007; 133:24-33; PMID:17631127; http://dx.doi.org/ 10.1053/j.gastro.2007.04.005 [DOI] [PubMed] [Google Scholar]
- [17].Cuoco L, Salvagnini M. Small intestine bacterial overgrowth in irritable bowel syndrome: a retrospective study with rifaximin. Minerva Gastroenterol Dietol 2006; 52:89-95; PMID:16554709 [PubMed] [Google Scholar]
- [18].Peralta S, Cottone C, Doveri T, Almasio PL, Craxi A. Small intestine bacterial overgrowth and irritable bowel syndrome-related symptoms: experience with Rifaximin. World J Gastroenterol 2009; 15:2628-31; PMID:19496193; http://dx.doi.org/ 10.3748/wjg.15.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yang J, Lee HR, Low K, Chatterjee S, Pimentel M. Rifaximin vs. other antibiotics in the primary treatment and retreatment of bacterial overgrowth in IBS. Dig Dis Sci 2008; 53:169-74; PMID:17520365; http://dx.doi.org/ 10.1007/s10620-007-9839-8 [DOI] [PubMed] [Google Scholar]
- [20].Lauritano EC, Gabrielli M, Lupascu A, Santoliquido A, Nucera G, Scarpellini E, Vincenti F, Cammarota G, Flore R, Pola P, et al.. Rifaximin dose-finding study for the treatment of small intestinal bacterial overgrowth. Aliment Pharmacol Ther 2005; 22:31-5; PMID:15963077; http://dx.doi.org/ 10.1111/j.1365-2036.2005.02516.x [DOI] [PubMed] [Google Scholar]
- [21].Lauritano EC, Gabrielli M, Scarpellini E, Ojetti V, Roccarina D, Villita A, Fiore E, Flore R, Santoliquido A, Tondi P, et al.. Antibiotic therapy in small intestinal bacterial overgrowth: rifaximin versus metronidazole. Eur Rev Med Pharmacol Sci 2009; 13:111-6; PMID:19499846 [PubMed] [Google Scholar]
- [22].Scarpellini E, Gabrielli M, Lauritano CE, Lupascu A, Merra G, Cammarota G, Cazzato IA, Gasbarrini G, Gasbarrini A. High dosage rifaximin for the treatment of small intestinal bacterial overgrowth. Aliment Pharmacol Ther 2007; 25:781-6; PMID:17373916; http://dx.doi.org/ 10.1111/j.1365-2036.2007.03259.x [DOI] [PubMed] [Google Scholar]
- [23].Pimentel M, Lembo A, Chey WD, Zakko S, Ringel Y, Yu J, Mareya SM, Shaw AL, Bortey E, Forbes WP, et al.. Rifaximin therapy for patients with irritable bowel syndrome without constipation. N Engl J Med 2011; 364:22-32; PMID:21208106; http://dx.doi.org/ 10.1056/NEJMoa1004409 [DOI] [PubMed] [Google Scholar]
- [24].Scarpellini E, Giorgio V, Gabrielli M, Filoni S, Vitale G, Tortora A, Ojetti V, Gigante G, Fundar∫ C, Gasbarrini A. Rifaximin treatment for small intestinal bacterial overgrowth in children with irritable bowel syndrome. Eur Rev Med Pharmacol Sci 2013; 17:1314-20; PMID:23740443 [PubMed] [Google Scholar]
- [25].Menees SB, Maneerattannaporn M, Kim HM, Chey WD. The efficacy and safety of rifaximin for the irritable bowel syndrome: a systematic review and meta-analysis. Am J Gastroenterol 2012; 107:28-35; quiz 36; PMID:22045120; http://dx.doi.org/ 10.1038/ajg.2011.355 [DOI] [PubMed] [Google Scholar]
- [26].Di Stefano M, Strocchi A, Malservisi S, Veneto G, Ferrieri A, Corazza GR. Non-absorbable antibiotics for managing intestinal gas production and gas-related symptoms. Aliment Pharmacol Ther 2000; 14:1001-8; PMID:10930893; http://dx.doi.org/ 10.1046/j.1365-2036.2000.00808.x [DOI] [PubMed] [Google Scholar]
- [27].Rezaie A, Nikfar S, Abdollahi M. The place of antibiotics in management of irritable bowel syndrome: a systematic review and meta-analysis. Arch Med Sci 2010; 6:49-55; PMID:22371720; http://dx.doi.org/ 10.5114/aoms.2010.13507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pimentel M. Review article: potential mechanisms of action of rifaximin in the management of irritable bowel syndrome with diarrhoea. Aliment Pharmacol Ther 2016; 43 Suppl 1:37-49; PMID:26618924; http://dx.doi.org/ 10.1111/apt.13437 [DOI] [PubMed] [Google Scholar]
- [29].Iorio N, Malik Z, Schey R. Profile of rifaximin and its potential in the treatment of irritable bowel syndrome. Clin Exp Gastroenterol 2015; 8:159-67; PMID:26089696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Meyrat P, Safroneeva E, Schoepfer AM. Rifaximin treatment for the irritable bowel syndrome with a positive lactulose hydrogen breath test improves symptoms for at least 3 months. Aliment Pharmacol Ther 2012; 36:1084-93; PMID:23066911; http://dx.doi.org/ 10.1111/apt.12087 [DOI] [PubMed] [Google Scholar]
- [31].Pimentel M, Park S, Mirocha J, Kane SV, Kong Y. The effect of a nonabsorbed oral antibiotic (rifaximin) on the symptoms of the irritable bowel syndrome: a randomized trial. Ann Intern Med 2006; 145:557-63; PMID:17043337; http://dx.doi.org/ 10.7326/0003-4819-145-8-200610170-00004 [DOI] [PubMed] [Google Scholar]
- [32].Sharara AI, Aoun E, Abdul-Baki H, Mounzer R, Sidani S, Elhajj I. A randomized double-blind placebo-controlled trial of rifaximin in patients with abdominal bloating and flatulence. Am J Gastroenterol 2006; 101:326-33; PMID:16454838; http://dx.doi.org/ 10.1111/j.1572-0241.2006.00458.x [DOI] [PubMed] [Google Scholar]
- [33].Jeffery IB, O'Toole PW, Öhman L, Claesson MJ, Deane J, Quigley EM, Simrén M. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 2012; 61:997-1006; PMID:22180058; http://dx.doi.org/ 10.1136/gutjnl-2011-301501 [DOI] [PubMed] [Google Scholar]
- [34].Soldi S, Vasileiadis S, Uggeri F, Campanale M, Morelli L, Fogli MV, Calanni F, Grimaldi M, Gasbarrini A. Modulation of the gut microbiota composition by rifaximin in non-constipated irritable bowel syndrome patients: a molecular approach. Clin Exp Gastroenterol 2015; 8:309-25; PMID:26673000; http://dx.doi.org/ 10.2147/CEG.S89999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Soares RL. Irritable bowel syndrome: a clinical review. World J Gastroenterol 2014; 20:12144-60; PMID:25232249; http://dx.doi.org/ 10.3748/wjg.v20.i34.12144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bellini M, Gambaccini D, Stasi C, Urbano MT, Marchi S, Usai-Satta P. Irritable bowel syndrome: a disease still searching for pathogenesis, diagnosis and therapy. World J Gastroenterol 2014; 20:8807-20; PMID:25083055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Xu D, Gao J, Gillilland M, Wu X, Song I, Kao JY, Owyang C. Rifaximin alters intestinal bacteria and prevents stress-induced gut inflammation and visceral hyperalgesia in rats. Gastroenterol 2014; 146:484-496. e4; PMID:24161699; http://dx.doi.org/ 10.1053/j.gastro.2013.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Le Gall G, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, Colquhoun IJ, Kemsley EK, Narbad A. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res 2011; 10:4208-18; PMID:21761941; http://dx.doi.org/ 10.1021/pr2003598 [DOI] [PubMed] [Google Scholar]
- [39].van Nuenen MH, de Ligt RA, Doornbos RP, van der Woude JC, Kuipers EJ, Venema K. The influence of microbial metabolites on human intestinal epithelial cells and macrophages in vitro. FEMS Immunol Med Microbiol 2005; 45:183-9; PMID:15939578; http://dx.doi.org/ 10.1016/j.femsim.2005.03.010 [DOI] [PubMed] [Google Scholar]
- [40].De Preter V, Verbeke K. Metabolomics as a diagnostic tool in gastroenterology. World J Gastrointest Pharmacol Ther 2013; 4:97-107; PMID:24199025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kajander K, Myllyluoma E, Kyrönpalo S, Rasmussen M, Sipponen P, Mattila I, Seppänen-Laakso T, Vapaatalo H, Oresic M, Korpela R. Elevated pro-inflammatory and lipotoxic mucosal lipids characterise irritable bowel syndrome. World J Gastroenterol 2009; 15:6068-74; PMID:20027679; http://dx.doi.org/ 10.3748/wjg.15.6068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ahmed I, Greenwood R, Costello Bde L, Ratcliffe NM, Probert CS. An investigation of fecal volatile organic metabolites in irritable bowel syndrome. PLoS One 2013; 8:e58204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ponnusamy K, Choi JN, Kim J, Lee SY, Lee CH. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J Med Microbiol 2011; 60:817-27; PMID:21330412; http://dx.doi.org/ 10.1099/jmm.0.028126-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zeber-Lubecka N, Kulecka M, Ambrozkiewicz F, Paziewska A, Lechowicz M, Konopka E, Majewska U, Borszewska-Kornacka M, Mikula M, Cukrowska B, et al.. Effect of Saccharomyces boulardii and Mode of Delivery on the Early Development of the Gut Microbial Community in Preterm Infants. PLoS One 2016; 11:e0150306; PMID:26918330; http://dx.doi.org/ 10.1371/journal.pone.0150306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al.. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009; 75:7537-41; PMID:19801464; http://dx.doi.org/ 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol 2012; 9:599-608; PMID:22907164; http://dx.doi.org/ 10.1038/nrgastro.2012.152 [DOI] [PubMed] [Google Scholar]
- [47].Morgan XC, Segata N, Huttenhower C. Biodiversity and functional genomics in the human microbiome. Trends Genet 2013; 29:51-8; PMID:23140990; http://dx.doi.org/ 10.1016/j.tig.2012.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al.. Enterotypes of the human gut microbiome. Nature 2011; 473:174-80; PMID:21508958; http://dx.doi.org/ 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Power SE, O'Toole PW, Stanton C, Ross RP, Fitzgerald GF. Intestinal microbiota, diet and health. Br J Nutr 2014; 111:387-402; PMID:23931069; http://dx.doi.org/ 10.1017/S0007114513002560 [DOI] [PubMed] [Google Scholar]
- [50].Ghorbani S, Nejad A, Law D, Chua KS, Amichai MM, Pimentel M. Healthy control subjects are poorly defined in case-control studies of irritable bowel syndrome. Ann Gastroenterol 2015; 28:87-93; PMID:25609236 [PMC free article] [PubMed] [Google Scholar]
- [51].Salipante SJ, Kawashima T, Rosenthal C, Hoogestraat DR, Cummings LA, Sengupta DJ, Harkins TT, Cookson BT, Hoffman NG. Performance comparison of Illumina and ion torrent next-generation sequencing platforms for 16S rRNA-based bacterial community profiling. Appl Environ Microbiol 2014; 80:7583-91; PMID:25261520; http://dx.doi.org/ 10.1128/AEM.02206-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Walker AW, Martin JC, Scott P, Parkhill J, Flint HJ, Scott KP. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 2015; 3:26; PMID:26120470; http://dx.doi.org/ 10.1186/s40168-015-0087-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rajilić-Stojanović M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol 2007; 9:2125-36; PMID:17686012; http://dx.doi.org/ 10.1111/j.1462-2920.2007.01369.x [DOI] [PubMed] [Google Scholar]
- [54].Krogius-Kurikka L, Lyra A, Malinen E, Aarnikunnas J, Tuimala J, Paulin L, Mäkivuokko H, Kajander K, Palva A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol 2009; 9:95; PMID:20015409; http://dx.doi.org/ 10.1186/1471-230X-9-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil 2012; 24:521-30, e248; PMID:22339879; http://dx.doi.org/ 10.1111/j.1365-2982.2012.01891.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Enck P, Zimmermann K, Rusch K, Schwiertz A, Klosterhalfen S, Frick J-S. The effects of ageing on the colonic bacterial microflora in adults. Z Gastroenterol 2009; 47:653-8; PMID:19606407; http://dx.doi.org/ 10.1055/s-0028-1109055 [DOI] [PubMed] [Google Scholar]
- [57].Chassard C, Dapoigny M, Scott KP, Crouzet L, Del'homme C, Marquet P, Martin JC, Pickering G, Ardid D, Eschalier A, et al.. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment Pharmacol Ther 2012; 35:828-38; PMID:22315951; http://dx.doi.org/ 10.1111/j.1365-2036.2012.05007.x [DOI] [PubMed] [Google Scholar]
- [58].Carroll IM, Chang YH, Park J, Sartor RB, Ringel Y. Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog 2010; 2:19; PMID:21143915; http://dx.doi.org/ 10.1186/1757-4749-2-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sheikh Sajjadieh MR, Kuznetsova LV, Bojenko VB. Dysbiosis in ukrainian children with irritable bowel syndrome affected by natural radiation. Iran J Pediatr 2012; 22:364-8; PMID:23400266 [PMC free article] [PubMed] [Google Scholar]
- [60].Tana C, Umesaki Y, Imaoka A, Handa T, Kanazawa M, Fukudo S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil 2010; 22:512-9, e114-115; PMID:19903265 [DOI] [PubMed] [Google Scholar]
- [61].Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science 2005; 308:1635-8; PMID:15831718; http://dx.doi.org/ 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kerckhoffs AP, Ben-Amor K, Samsom M, van der Rest ME, de Vogel J, Knol J, Akkermans LM. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J Med Microbiol 2011; 60:236-45; PMID:20947663; http://dx.doi.org/ 10.1099/jmm.0.022848-0 [DOI] [PubMed] [Google Scholar]
- [63].Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, et al.. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 2011; 141:1782-91; PMID:21741921; http://dx.doi.org/ 10.1053/j.gastro.2011.06.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Rigsbee L, Agans R, Shankar V, Kenche H, Khamis HJ, Michail S, Paliy O. Quantitative profiling of gut microbiota of children with diarrhea-predominant irritable bowel syndrome. Am J Gastroenterol 2012; 107:1740-51; PMID:22986438; http://dx.doi.org/ 10.1038/ajg.2012.287 [DOI] [PubMed] [Google Scholar]
- [65].Durbán A, Abellán JJ, Jiménez-Hernández N, Salgado P, Ponce M, Ponce J, Garrigues V, Latorre A, Moya A. Structural alterations of faecal and mucosa-associated bacterial communities in irritable bowel syndrome. Environ Microbiol Rep 2012; 4:242-7; http://dx.doi.org/ 10.1111/j.1758-2229.2012.00327.x [DOI] [PubMed] [Google Scholar]
- [66].Spiller R, Garsed K. Postinfectious irritable bowel syndrome. Gastroenterology 2009; 136:1979-88; PMID:19457422; http://dx.doi.org/ 10.1053/j.gastro.2009.02.074 [DOI] [PubMed] [Google Scholar]
- [67].Pimentel M, Lembo A, Chey WD, Zakko S, Ringel Y, Yu J, Mareya SM, Shaw AL, Bortey E, Forbes WP, et al.. Rifaximin therapy for patients with irritable bowel syndrome without constipation. N Engl J Med 2011; 364:22-32; PMID:21208106; http://dx.doi.org/ 10.1056/NEJMoa1004409 [DOI] [PubMed] [Google Scholar]
- [68].Wall GC, Bryant GA, Bottenberg MM, Maki ED, Miesner AR. Irritable bowel syndrome: a concise review of current treatment concepts. World J Gastroenterol 2014; 20:8796-806; PMID:25083054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Schoenfeld P, Pimentel M, Chang L, Lembo A, Chey WD, Yu J, Paterson C, Bortey E, Forbes WP. Safety and tolerability of rifaximin for the treatment of irritable bowel syndrome without constipation: a pooled analysis of randomised, double-blind, placebo-controlled trials. Aliment Pharmacol Ther 2014; 39:1161-8; PMID:24697851; http://dx.doi.org/ 10.1111/apt.12735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Saadi M, McCallum RW. Rifaximin in irritable bowel syndrome: rationale, evidence and clinical use. Ther Adv Chronic Dis 2013; 4:71-5; PMID:23556126; http://dx.doi.org/ 10.1177/2040622312472008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Koo HL, Sabounchi S, Huang DB, DuPont HL. Rifaximin therapy of irritable bowel syndrome. Clin Med Insights Gastroenterol 2012; 5:31-41; PMID:24833932; http://dx.doi.org/ 10.4137/CGast.S7382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Frissora CL, Cash BD. Review article: the role of antibiotics vs. conventional pharmacotherapy in treating symptoms of irritable bowel syndrome. Aliment Pharmacol Ther 2007; 25:1271-81; PMID:17509095; http://dx.doi.org/ 10.1111/j.1365-2036.2007.03313.x [DOI] [PubMed] [Google Scholar]
- [73].Bajaj JS, Heuman DM, Sanyal AJ, Hylemon PB, Sterling RK, Stravitz RT, Fuchs M, Ridlon JM, Daita K, Monteith P, et al.. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS One 2013; 8:e60042; PMID:23565181; http://dx.doi.org/ 10.1371/journal.pone.0060042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, et al.. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol 2013; 58:949-55; PMID:23333527; http://dx.doi.org/ 10.1016/j.jhep.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Monleón D, Morales JM, Barrasa A, López JA, Vázquez C, Celda B. Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed 2009; 22:342-8; http://dx.doi.org/ 10.1002/nbm.1345 [DOI] [PubMed] [Google Scholar]
- [76].Elia M, Engfer MB, Green CJ, Silk DB. Systematic review and meta-analysis: the clinical and physiological effects of fibre-containing enteral formulae. Aliment Pharmacol Ther 2008; 27:120-45; PMID:17922802; http://dx.doi.org/ 10.1111/j.1365-2036.2007.03544.x [DOI] [PubMed] [Google Scholar]
- [77].Zumarraga L, Levitt MD, Suarez F. Absence of gaseous symptoms during ingestion of commercial fibre preparations. Aliment Pharmacol Ther 1997; 11:1067-72; PMID:9663831; http://dx.doi.org/ 10.1046/j.1365-2036.1997.00250.x [DOI] [PubMed] [Google Scholar]
- [78].Dlugosz A, Winckler B, Lundin E, Zakikhany K, Sandström G, Ye W, Engstrand L, Lindberg G. No difference in small bowel microbiota between patients with irritable bowel syndrome and healthy controls. Sci Rep 2015; 5:8508; PMID:25687743; http://dx.doi.org/ 10.1038/srep08508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology 2006; 130:1480-91; PMID:16678561; http://dx.doi.org/ 10.1053/j.gastro.2005.11.061 [DOI] [PubMed] [Google Scholar]
- [80].Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078-9; PMID:19505943; http://dx.doi.org/ 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].FASTX-Toolkit [Internet]. [cited 2015 Apr 6]; Available from: http://hannonlab.cshl.edu/fastx_toolkit/index.html [Google Scholar]
- [82].Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 2014; 30:3123-4; PMID:25061070; http://dx.doi.org/ 10.1093/bioinformatics/btu494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wickham H. ggplot2: Elegant Graphics for Data Analysis 1st ed. 2009. Corr. 3rd printing 2010 edition. New York: Springer; 2010 [Google Scholar]
- [84].Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Statistical Society Series B (Methodological) 1995; 57:289-300. [Google Scholar]
- [85].Faul F, Erdfelder E, Lang A-G, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 2007; 39:175-91; PMID:17695343; http://dx.doi.org/ 10.3758/BF03193146 [DOI] [PubMed] [Google Scholar]
- [86].Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al.. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013; 31:814-21; PMID:23975157; http://dx.doi.org/ 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Dawiskiba T, Deja S, Mulak A, Ząbek A, Jawień E, Pawełka D, Banasik M, Mastalerz-Migas A, Balcerzak W, Kaliszewski K, et al.. Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J Gastroenterol 2014; 20:163-74; PMID:24415869; http://dx.doi.org/ 10.3748/wjg.v20.i1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kennard RW, Stone LA. Computer Aided Design of Experiments. Technometrics 1969; 11:137-48; http://dx.doi.org/ 10.1080/00401706.1969.10490666 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.