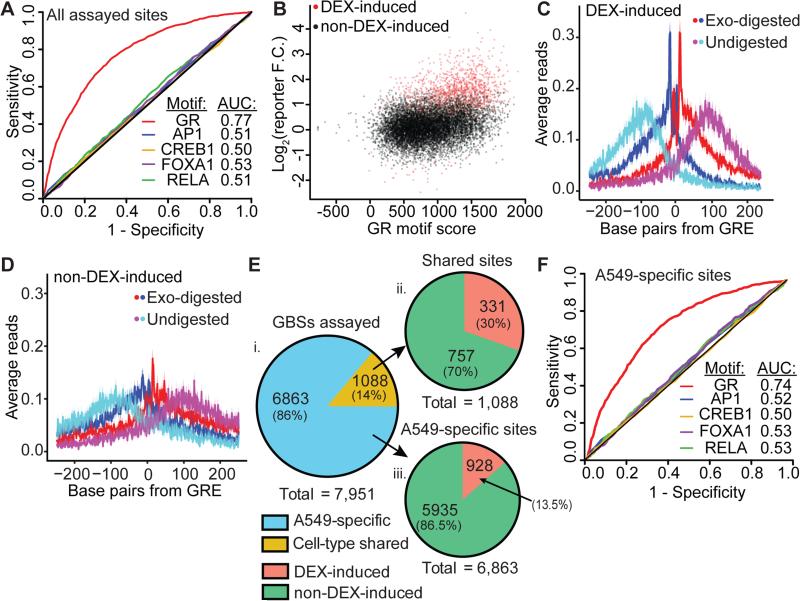
Figure 2. GRE-encoded GBSs confer DEX-inducible enhancer function.
(A) ROC analysis of DEX-induced enhancers evaluating DNA sequence motif significance for the GRE and known GR interacting proteins as predictors of enhancer function. (B) Scatter plot of GR motif score vs. ChIP-reporter activity. Significant DEX-induced elements are shown in red (r = 0.35). (C) Average GR ChIP-exo signal across DEX-inducible enhancer GBSs centered on the strongest GRE. Red and blue are the 5’ bp of reads aligned to the positive or negative strand, respectively. Cyan and magenta are the 5’ bp of reads at non-exonuclease treated fragment end aligned to the positive or negative strand, respectively. Ribbons indicate standard error. (D) GR ChIP-exo signal as above, aggregated across an equal number of sub-sampled GBSs that are not induced by DEX in reporter assays. (E) i. Fraction of GBSs in a previous study assayed using ChIP-reporters that were specific to A549 cells or common between A549 and Ishikawa cells (N = 7,771). ii. Fraction of A549-Ishikawa shared GBSs with significant DEX-induced reporter activity (N = 1088). iii. Fraction of A549-specific sites with significant DEX-induced reporter activity (N = 6,863). GBSs are categorized as specific to A549 cells or shared between A549 cells and endometrium-derived Ishikawa cells (Gertz et al. 2013). (F) ROC analysis of A549-specific DEX-inducible enhancers.
See also Figure S3.