Abstract
Background
The response of rectal cancers to neoadjuvant chemoradiation (CRT) is variable, but tools to predict response remain lacking. We evaluated whether KRAS and TP53 mutations are associated with pathologic complete response (pCR) and lymph node metastasis after adjusting for neoadjuvant regimen.
Methods
Retrospective analysis of 229 pretreatment biopsies from patients with stage II/III rectal cancer was performed. All patients received CRT. Patients received zero to eight cycles of FOLFOX either before or after CRT, but prior to surgical excision. A subset was analyzed to assess concordance between mutation calls by Sanger sequencing and a next-generation assay.
Results
96 (42%) tumors had KRAS mutation, 150 had TP53 mutation (66%), and 59 (26%) had both. 59 patients (26%) achieved pCR following neoadjuvant therapy. 45 of 133 (34%) KRAS wild-type tumors had pCR, compared with 14 of 96 (15%) KRAS mutant tumors (p=0.001). KRAS mutation remained independently associated with a lower pCR rate on multivariable analysis after adjusting for clinical stage, CRT-to-surgery interval, and cycles of FOLFOX (OR 0.34, 95% CI: 0.17-0.66, p < 0.01). Of 29 patients with KRAS G12V or G13D, only 2 (7%) achieved pCR. Tumors with both KRAS and TP53 mutation were associated with lymph node metastasis. The concordance between platforms was high for KRAS (40 of 43, 93%).
Conclusions
KRAS mutation is independently associated with a lower pCR rate in locally advanced rectal cancer after adjusting for variations in neoadjuvant regimen. Genomic data can potentially be used to select patients for “watch and wait” strategies.
Background
The response of locally advanced rectal cancer (LARC) to neoadjuvant chemoradiation (CRT) is variable. Some tumors respond completely, achieving pathologic complete response (pCR), while others have very limited response. For tumors that show pCR, the role of radical surgery has been called into question and “watch and wait” approaches are being explored.1,2 For unresponsive tumors, surgical resection remains crucial to minimize risk of progression.
Because there are no validated tools to predict pCR for patients prior to surgical resection, offering “watch and wait” or limited resections seems risky as it might compromise oncologic efficacy. Digital rectal examination, CT/PET-CT, and MRI have all been shown to be relatively unreliable at predicting pCR, with accuracies ranging from 40%-80%.3–7 If, however, we could identify patients likely to achieve pCR in advance, we could spare them radical surgical excision with its associated morbidities. Similarly, if we could identify patients unlikely to have lymph node metastases, we might isolate candidates for whom a limited resection could be sufficient.
We previously reported that tumors with KRAS mutation and those with both KRAS and TP53 mutations were associated with resistance to CRT based on an early subset of the trial-derived specimens included in the current analysis. At that time, our cohort was not positioned to account for differences in the neoadjuvant regimen that were inherent in the study protocol.8 Since then, the phase 2 trial concluded, and we learned that increasing the duration between CRT and surgery by administering additional cycles of FOLFOX could increase the pCR rate from 19% (when CRT was followed by surgery after 6-8 weeks) to 38% (when CRT was followed by up to 6 cycles of FOLFOX, which concurrently increased the median CRT-to-surgery interval to 19 weeks).9 This finding was consistent with other retrospective reports that identified higher rates of tumor response with prolonged intervals between CRT and surgery.10–13 Other groups have reported response rates over 30% by incorporating systemic chemotherapy prior to chemoradiation therapy.14,15
In this study, we corroborate our previous finding that tumors with KRAS mutation and combined KRAS/TP53 mutations are associated with resistance to neoadjuvant therapy using a larger, expanded cohort. We show that this association persists after accounting for other factors that influence response including stage, the number of cycles of neoadjuvant FOLFOX, and the CRT-to-surgery interval by leveraging the heterogeneity in neoadjuvant regimens within this expanded cohort. We also show that tumors with both KRAS and TP53 mutations are associated with a higher rate of lymph node metastasis. Finally, we demonstrate a high concordance between KRAS mutation calls by Sanger Sequencing and a hybridization capture-based next-generation sequencing assay.
Methods
Study Population
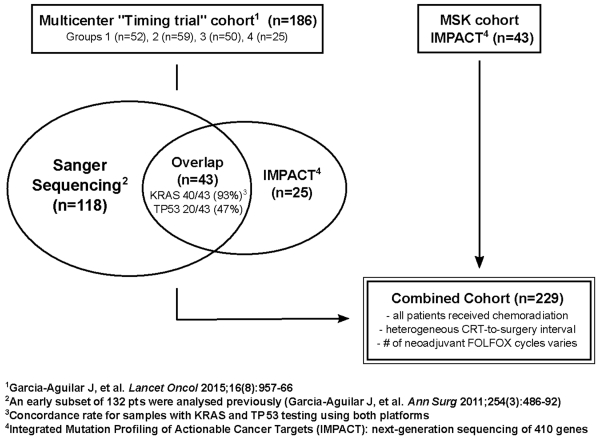
Tissue from patients in the “Timing trial”, a multi-center, prospective, phase 2 clinical trial (ClinicalTrials.gov, number NCT00335816), and from patients treated at Memorial Sloan Kettering Cancer Center (MSK), were combined to create a cohort of LARC treated with neoadjuvant regimens of various intensities (Figure 1). All patients had American Joint Committee on Cancer (AJCC) clinical stage II (T3-4, N0) or III (any T, N1-2) rectal adenocarcinoma with a distal tumor border within 15 cm of the anal verge by proctoscopy. Local staging was performed by endorectal ultrasound or MRI, and patients were screened for metastatic disease with CT. Inclusion was also contingent on an adequate amount of tissue in pre-treatment diagnostic biopsies to allow for mutational profiling. The use of specimens for molecular analysis was approved by the Institutional Review Board (IRB) at each institution, and consent was obtained for the use of tissue specimens.
Figure 1. Study Cohort Composition.
Sample Preparation and KRAS testing
Pretreatment biopsies were formalin-fixed, paraffin-embedded, and placed on slides that were marked by a pathologist and manually microdissected for tumor tissue. Genomic DNA was extracted using QIAamp or AllPrep DNA FFPE Tissue kits (Qiagen Inc., Valencia, CA). KRAS and TP53 mutations were originally determined by standard polymerase chain reaction (PCR) followed by Sanger Sequencing of exons 2 and 3 of KRAS or exons 4-8 and the Pro72 sequence of TP53 (Supplementary Table 1). Recently, we transitioned to a hybridization capture-based next-generation sequencing assay, MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets), which sequences all exons and selected introns of 410 cancer genes, including KRAS and TP53.16,17 A subset of the combined cohort was analyzed by both, allowing us to assess concordance between techniques. When discordance was noted for samples analyzed by both platforms, we favored the call made by MSK-IMPACT as it included coding sequences not included by Sanger Sequencing.
Treatment Received and Pathologic Assessment
All patients received neoadjuvant radiotherapy with a fluoropyrimidine-based chemosensitizing agent. The patients from the “Timing trial” fall into 4 groups corresponding to the trial protocol, with patients receiving CRT followed by 0, 2, 4, or 6 cycles of FOLFOX before surgery. Patients treated at MSK generally received up to 8 cycles of FOLFOX prior to CRT. Following neoadjuvant therapy, surgical resection was performed using the principles of sharp total mesorectal excision. Pathologic complete response was defined as the absence of tumor cells in the surgical specimen at the primary tumor site and regional lymph nodes.
Statistical Considerations
Retrospective analysis was performed based on treatment-received. The testing of patient characteristics comparing those with pCR vs. non-pCR, and the presence vs. absence of lymph node metastasis on pathology, were performed using Fisher’s test for categorical variables and Wilcoxon rank sum test for continuous variables. Multivariable analysis was performed adjusting for variables that were either significant in univariable analysis or those widely considered to be relevant prognostic factors.
Results
Patient and Treatment Characteristics
A total of 229 patients were included (Figure 1); 186 patients (81%) were enrolled in the “Timing trial”, and 43 (19%) from the MSK cohort. Of the patients from the “Timing trial”, 132 were previously analyzed.8 Patients in the “Timing trial” received 0-6 cycles FOLFOX after CRT and had surgery between 5.4-61.4 weeks after CRT completion. Patients in the MSK cohort received between 0–8 cycles FOLFOX cycles before CRT, and had surgery between 4.6–23.1 weeks after completion of CRT.
KRAS and TP53 mutation calls and concordance between platforms
In total, 161 of the 229 patients (70%) had KRAS and TP53 mutation status testing of their biopsies by Sanger Sequencing, while 111 patients (48%) were evaluated by MSK-IMPACT. A subset of 43 patients was analyzed using both platforms, and the concordance rate for KRAS mutation calls was 40 out of 43 (93%).
One patient with a KRAS A146P mutation (exon 4) recognized by MSK-IMPACT was misclassified as wild-type by Sanger Sequencing because only exons 2 and 3 were evaluated. Another patient classified as wild-type by Sanger Sequencing was found to have a G12S mutation that was captured by MSK-IMPACT. The variant frequency was only 2.4% in the sample processed by MSK-IMPACT, making it unlikely that it could have been identified by standard Sanger Sequencing methodology. The last patient with a discordant KRAS call had G13D mutation by Sanger Sequencing that was not recognized by MSK-IMPACT, and was counted as wild-type after further group review of the sequencing data. Among patients sequenced by both platforms, TP53 mutations were identified by MSK-IMPACT in 32 (74%), whereas Sanger Sequencing only identified 18 (42%), with concordance between platforms in only 20 out of 43 (47%).
KRAS mutation and pCR
In the entire combined cohort of 229 patients, 14 of 96 (15%) patients with KRAS mutations achieved pCR, whereas 45 of 133 (34%) patients with KRAS wild-type tumors achieved pCR (p = 0.001, Table 1). This result expands on, and closely corroborates, our report on the first 132 patients of the “Timing trial”, where 14% of patients with KRAS mutation achieved pCR compared with 33% of those with wild-type KRAS.8 Tumors with both KRAS and TP53 mutation were associated with a particularly low pCR rate (6 of 59, 10%), which also corroborated our prior claim that the double-mutants may be associated with particularly low rates of response.
Table 1. Patient Characteristics and Univariable Association with pCR.
| Variable | Total (n=229) |
Non-pCR (n=170) |
pCR (n=59) |
p-value |
|---|---|---|---|---|
| Age at Diagnosis | 56 (21 - 87) | 56.5 (21 - 87) | 55 (32 - 80) | 0.499 |
| Sex | ||||
| Female | 94 (41) | 68 (72) | 26 (28) | 0.646 |
| Male | 135 (59) | 102 (76) | 33 (24) | |
| Race | ||||
| Asian | 13 (6) | 9 (69) | 4 (31) | 0.190 |
| Black | 9 (4) | 4 (44) | 5 (56) | |
| White | 192 (84) | 145 (76) | 47 (24) | |
| Unknown | 15 (7) | 12 (80) | 3 (20) | |
| Clinical Stage | ||||
| II | 52 (23) | 40 (77) | 12 (23) | 0.719 |
| III | 177 (77) | 130 (73) | 47 (27) | |
| Time between CRT and Surgery | ||||
| < 8 Weeks | 65 (28) | 53 (82) | 12 (18) | 0.133 |
| 8+ Weeks | 164 (72) | 117 (71) | 47 (29) | |
| Cycles of FOLFOX | ||||
| < 3 cycles | 135 (59) | 103 (76) | 32 (24) | 0.443 |
| 3+ cycles | 94 (41) | 67 (71) | 27 (29) | |
| KRAS | ||||
| Wild-type | 133 (58) | 88 (66) | 45 (34) | 0.001 |
| Mutant | 96 (42) | 82 (85) | 14 (15) | |
| TP53 | ||||
| Wild-type | 79 (34) | 58 (73) | 21 (27) | 0.874 |
| Mutant | 150 (66) | 112 (75) | 38 (25) | |
| KRAS and TP53 | ||||
| Not double-mutant | 170 (74) | 117 (69) | 53 (31) | 0.001 |
| Double mutant | 59 (26) | 53 (90) | 6 (10) |
Median (range) are presented for continuous variables, count (percentage) for categorical variables. Testing of patient characteristics between those with pCR and without pCR uses Fisher’s test for categorical variables and Wilcoxon rank sum test for continuous variables. pCR – pathologic complete response.
Univariable and Multivariable Analysis of Clinical and Treatment-related Characteristics and their Association with pCR
Because previous work suggests a longer CRT-to-surgery interval and more cycles of chemotherapy provided in the neoadjuvant setting are associated with increased rate of pCR, we wanted to test whether KRAS mutation was independently associated with pCR in this combined cohort after controlling for these factors.
Tumors with KRAS mutation, and also those with both KRAS and TP53 mutations were negatively associated with pCR (both p = 0.001, Table 1). On multivariable analysis, KRAS mutation remained independently associated with pCR with an odds ratio of 0.34 (95% CI: 0.17-0.66, p = 0.002) after adjusting for clinical stage, time from CRT-to-surgery, and cycles of FOLFOX received (Table 2). Interestingly, the CRT-to-surgery interval and whether the patient received greater or less than 3 cycles of FOLFOX in the neoadjuvant setting were not significantly associated with pCR in this cohort.
Table 2. Multivariable Analysis for Association with pCR.
| Variables | OR | 95% CI | p-value |
|---|---|---|---|
| KRAS (Mutant vs. Wild-type) | 0.34 | (0.17 - 0.66) | 0.002 |
| Clinical Stage (III vs. II) | 1.23 | (0.58 - 2.60) | 0.585 |
| Time from CRT to Surgery (8+ weeks vs. < 8 weeks) | 1.70 | (0.80 - 3.62) | 0.171 |
| Cycles of FOLFOX (3+ vs. < 3) | 1.09 | (0.58 - 2.07) | 0.788 |
Collectively, these data indicate that the KRAS genotype of rectal cancer is associated with a decreased incidence of pCR, independent of other treatment related variables such as the CRT-to-surgery interval and the number of cycles of FOLFOX. The combination of KRAS and TP53 mutations was not included in the multivariable analysis because it is collinear with KRAS mutation.
KRAS G12V and G13D Mutations are Associated with Lower pCR
It has previously been reported that the specific KRAS mutation may result in variable responsiveness to CRT.18 In this cohort, out of 14 patients with G12V mutation, only one (7%) achieved pCR, and of 15 patients with G13D mutation, only one (7%) achieved pCR (Table 3). This corroborates our prior observation that rectal cancers with mutations in codon 13 and the G12V variant of KRAS appear particularly resistant to neoadjuvant therapy. In contrast, 8 of 41 patients (20%) with G12D mutations achieved pCR.
Table 3. Distribution of KRAS mutations and number that achieved pCR.
(% achieving pCR included in parentheses for KRAS mutations that were identified in more than 10 samples)
| KRAS Mutation | n | n pCR | |
|---|---|---|---|
|
| |||
| Codon 12 | G12D | 41 | 8 (20%) |
| G12V | 14 | 1 (7%) | |
| G12S | 4 | 0 | |
| G12A | 4 | 0 | |
| G12C | 2 | 1 | |
| G12R | 1 | 1 | |
| Codon 13 | G13D | 15 | 1 (7%) |
| G13C | 1 | 0 | |
| Codon 61 | Q61L | 3 | 1 |
| Q61H | 2 | 0 | |
| del 2 or 4bp | 2 | 0 | |
| Codon 146 | A146T | 2 | 0 |
| A146P | 1 | 0 | |
| Codon 6 | L6F | 1 | 0 |
| Codon 34 | P34R | 1 | 1 |
| Codon 64 | Y64H | 1 | 0 |
| Codon 117 | K117N | 1 | 0 |
|
| |||
| Total | 96 | 14 | |
Mutation Profile and Lymph Node Metastasis
The presence of lymph node metastasis is clinically relevant because it reflects an increased risk for distant progression. Furthermore, if lymph node involvement is found in a resected specimen, it follows that a local resection (e.g. transanal excision) would have been inadequate to achieve locoregional control. We therefore evaluated whether mutational profile was associated with lymph node metastasis, and found that younger age, the combination of KRAS and TP53 mutations, and receiving less than 3 cycles of FOLFOX were each associated with lymph node metastasis in the pathologic specimen on univariable analysis (p < 0.05, Supplementary Table 2). On multivariable analysis, after adjusting for pre-clinical stage, these variables remained independently associated with lymph node metastasis (Table 4).
Table 4. Multivariable Analysis for Association with Lymph Node Metastasis.
| Variables | OR | 95% CI | p-value |
|---|---|---|---|
| Age at Diagnosis | 0.97 | (0.94 - 1.00) | 0.024 |
| KRAS/TP53 (Double-mutant vs. Not double-mutant) | 2.58 | (1.24 - 5.34) | 0.011 |
| Clinical Stage (III vs. II) | 2.47 | (0.90 - 6.82) | 0.080 |
| Cycles of FOLFOX (3+ vs. < 3) | 0.40 | (0.19 - 0.86) | 0.018 |
Discussion
The ability to predict LARC response to neoadjuvant chemotherapy is a crucial step to determining which patients are most suitable for a “watch and wait” approach. Testing for KRAS mutation has been successfully incorporated into the treatment algorithm for metastatic colorectal cancer to identify patients suitable for anti-EGFR therapy.19 To our knowledge, this is the first study evaluating KRAS and TP53 mutation with respect to LARC response to neoadjuvant therapy that takes into account the potential confounders of CRT-to-surgery interval, cycles of chemotherapy, and pre-treatment clinical stage. It also represents the largest cohort analyzed for an association between KRAS and TP53 mutations and pCR or lymph node metastasis in LARC. Our findings suggest a role for KRAS testing in patients with LARC to predict response to neoadjuvant therapy. Since tumors with KRAS mutation or both KRAS and TP53 mutations are less likely to achieve complete response to neoadjuvant therapy, they may be better suited for standard surgical resection. Knowing the KRAS status of tumors may ultimately guide decision making for patients who have tumors that appear to respond completely (or almost completely) to neoadjuvant therapy.
KRAS mutation has been studied as a potential prognostic marker for patients with colorectal cancer for over a quarter-century.20 Extensive research on the MAPK pathway and its regulation of cell proliferation led to our current understanding of the selective benefit of anti-EGFR therapies for patients with wild-type KRAS/NRAS.21 Our finding that KRAS mutation was independently associated with lower rates of complete response despite the fact that no targeted agents were involved in the neoadjuvant regimens for these patients is striking. It suggests that KRAS mutation in LARC has prognostic implications beyond its known relevance to resistance of anti-EGFR therapies, and provides a basis for further hypotheses to explore both clinically and within the laboratory. This finding corresponds with observations that particular KRAS mutations are associated with poor response to adjuvant FOLFOX in advanced colon cancers, though the mechanisms have not been elucidated.22,23 It is worth noting that not all studies have found an association between KRAS and poor response of LARC to neoadjuvant therapy, but most published studies to date have been small, retrospective cohorts that included tumors of varied clinical stage and neoadjuvant regimen.24 Our finding that KRAS and TP53 mutation are independently associated with lymph node metastasis is also striking as the presence of lymph node involvement reflects a higher risk of progression to distant metastasis. As such, patients with lymph node metastasis would be inadequately treated with local resection or “watch and wait”, and the combination of KRAS and TP53 mutation may reflect poor candidacy for these approaches.
The treatment-related variables had effects on response that were expected based on our trial results and the results of others. While the duration between CRT and surgery was not independently associated with pCR in this cohort, there remained a trend towards longer duration being associated with higher rate of pCR. The number of cycles of FOLFOX given in the neoadjuvant setting was found to be independently associated with lymph node metastases, with less cycles of neoadjuvant FOLFOX being associated with lymph node involvement in the resected specimen, which is both intuitive and biologically plausible.
Our study demonstrates that KRAS testing by either traditional Sanger Sequencing or a hybridization capture-based next-generation approach such as MSK-IMPACT was effective at identifying mutations with a high level of concordance. In our cohort, KRAS G12V and G13D mutations appeared to represent prevalent variants that were particularly resistant to neoadjuvant therapy and unlikely to achieve pCR. This finding corroborates our previous work and the observations of others.18,25
We noted a higher discordance rate between our two platforms in testing for TP53 mutations, that is likely due to the distribution of mutations across coding exons of TP53 that were not subjected to Sanger Sequencing. Despite this limitation, the association between tumors with both KRAS and TP53 mutations and lymph node metastasis remains provocative, suggesting that the genomic profile of an individual tumor may be able to delineate not only complete response from non-response, but also its propensity to metastasize to the lymph nodes.
This study has the advantages of including a large cohort of rectal cancer patients with pretreatment tissue, but has the limitations inherent to all retrospective studies. While the patient population is representative of the rectal cancer patients eligible for neoadjuvant therapy under current guidelines, the treatment regimens were heterogenous. Some patients received only CRT before surgery while others received a variable number of cycles of FOLFOX before or after the CRT but before surgery. This treatment heterogeneity is expected to have influenced the rate of response and therefore directly impacts the association of mutation status with pCR. On the other hand, we leveraged this heterogeneity to establish whether KRAS was independently associated with the rate of pCR after accounting for treatment-related variables. Because the chemoradiation regimen was standard and well-tolerated across nearly all patients in this cohort, we did not further specify the exact radiation dose and features in this study. Rather, we intentionally focused the assessment on the variation in systemic chemotherapeutic agents provided before or after chemoradiation, since this has been the focus of most clinical trial efforts presently. Another important limitation is that the majority of KRAS mutant variants did not occur with frequency. Indeed, only G12D, G12V, and G13D variants were present in more than 10 samples, but both G12V and G13D had particularly low rates of pCR in our cohort, which could prove useful in identifying tumors unlikely to achieve complete response.
In summary, patients with LARC treated with neoadjuvant therapy are less likely to have pCR when the tumor has a KRAS mutation, independent of other tumor or treatment characteristics. KRAS G12V and G13D appear particularly resistant to neoadjuvant therapy. These results suggest that genomic profiling can potentially be used to guide the selection of suitable candidates for “watch and wait” strategies after neoadjuvant therapy. Further investigational studies should prospectively evaluate the use of KRAS and TP53 testing to stratify patients to different therapeutic algorithms based on predicted response to neoadjuvant therapy.
Supplementary Material
Synopsis.
The response of rectal cancer to neoadjuvant therapy varies, but remains unpredictable. We analyzed 229 locally advanced rectal cancers treated with neoadjuvant therapy and found that tumors with KRAS mutation or both KRAS and TP53 mutations are independently associated with decreased rates of pathologic complete response.
Acknowledgments
This study was supported by the National Institutes of Health (NIH), National Cancer Institute (NCI) R01 Grant CA090559 (JGA).
Collaborators
Additional thanks for the contributions of Garrett Nash, Larissa Temple, José Guillem, Philip Paty (Surgical faculty members from the Colorectal Service of Memorial Sloan Kettering Cancer Center) and members of the Timing of Rectal Cancer Response to Chemoradiation Consortium: David Smith, Jorge Marcet, Peter Cataldo, Madhulika Varma, Anjali Kumar, Samuel Oommen, Theodore Coutsoftides, Steven Hunt, Michael Stamos, Charles Ternent, Daniel Herzig, Alessandro Fichera, Blase Polite, David Dietz.
References
- 1.Habr-Gama A, Perez RO, Nadalin W, et al. Operative versus nonoperative treatment for stage 0 distal rectal cancer following chemoradiation therapy: long-term results. Ann Surg. 2004;240(4):711–717. doi: 10.1097/01.sla.0000141194.27992.32. discussion 717-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JD, Ruby JA, Goodman KA, et al. Nonoperative management of rectal cancer with complete clinical response after neoadjuvant therapy. Ann Surg. 2012;256(6):965–972. doi: 10.1097/SLA.0b013e3182759f1c. doi:10.1097/SLA.0b013e3182759f1c. [DOI] [PubMed] [Google Scholar]
- 3.Guillem JG, Chessin DB, Shia J, et al. Clinical examination following preoperative chemoradiation for rectal cancer is not a reliable surrogate end point. J Clin Oncol. 2005;23(15):3475–3479. doi: 10.1200/JCO.2005.06.114. doi:10.1200/JCO.2005.06.114. [DOI] [PubMed] [Google Scholar]
- 4.Radovanovic Z, Breberina M, Petrovic T, Golubovic A, Radovanovic D. Accuracy of endorectal ultrasonography in staging locally advanced rectal cancer after preoperative chemoradiation. Surg Endosc. 2008;22(11):2412–2415. doi: 10.1007/s00464-008-0037-3. doi:10.1007/s00464-008-0037-3. [DOI] [PubMed] [Google Scholar]
- 5.Huh JW, Park YA, Jung EJ, Lee KY, Sohn S-K. Accuracy of endorectal ultrasonography and computed tomography for restaging rectal cancer after preoperative chemoradiation. J Am Coll Surg. 2008;207(1):7–12. doi: 10.1016/j.jamcollsurg.2008.01.002. doi:10.1016/j.jamcollsurg.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Leibold T, Akhurst TJ, Chessin DB, et al. Evaluation of 18F-FDG-PET for early detection of suboptimal response of rectal cancer to preoperative chemoradiotherapy: a prospective analysis. Ann Surg Oncol. 2011;18(10):2783–2789. doi: 10.1245/s10434-011-1634-2. doi:10.1245/s10434-011-1634-2. [DOI] [PubMed] [Google Scholar]
- 7.Memon S, Lynch AC, Bressel M, Wise AG, Heriot AG. Systematic review and meta-analysis of the accuracy of MRI and ERUS in the restaging and response assessment of rectal cancer following neoadjuvant therapy. Colorectal Dis. 2015 doi: 10.1111/codi.12976. doi:10.1111/codi.12976. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Aguilar J, Chen Z, Smith DD, et al. Identification of a biomarker profile associated with resistance to neoadjuvant chemoradiation therapy in rectal cancer. Ann Surg. 2011;254(3):486–492. doi: 10.1097/SLA.0b013e31822b8cfa. discussion 492-493. doi:10.1097/SLA.0b013e31822b8cfa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Aguilar J, Chow OS, Smith DD, et al. Effect of adding mFOLFOX6 after neoadjuvant chemoradiation in locally advanced rectal cancer: a multicentre, phase 2 trial. Lancet Oncol. 2015;16(8):957–966. doi: 10.1016/S1470-2045(15)00004-2. doi:10.1016/S1470-2045(15)00004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalady MF, de Campos-Lobato LF, Stocchi L, et al. Predictive factors of pathologic complete response after neoadjuvant chemoradiation for rectal cancer. Ann Surg. 2009;250(4):582–589. doi: 10.1097/SLA.0b013e3181b91e63. doi:10.1097/SLA.0b013e3181b91e63. [DOI] [PubMed] [Google Scholar]
- 11.Wolthuis AM, Penninckx F, Haustermans K, et al. Impact of interval between neoadjuvant chemoradiotherapy and TME for locally advanced rectal cancer on pathologic response and oncologic outcome. Ann Surg Oncol. 2012;19(9):2833–2841. doi: 10.1245/s10434-012-2327-1. doi:10.1245/s10434-012-2327-1. [DOI] [PubMed] [Google Scholar]
- 12.Zeng W-G, Zhou Z-X, Liang J-W, et al. Impact of interval between neoadjuvant chemoradiotherapy and surgery for rectal cancer on surgical and oncologic outcome. J Surg Oncol. 2014;110(4):463–467. doi: 10.1002/jso.23665. doi:10.1002/jso.23665. [DOI] [PubMed] [Google Scholar]
- 13.Calvo FA, Morillo V, Santos M, et al. Interval between neoadjuvant treatment and definitive surgery in locally advanced rectal cancer: impact on response and oncologic outcomes. J Cancer Res Clin Oncol. 2014;140(10):1651–1660. doi: 10.1007/s00432-014-1718-z. doi:10.1007/s00432-014-1718-z. [DOI] [PubMed] [Google Scholar]
- 14.Cercek A, Goodman KA, Hajj C, et al. [Accessed December 15, 2014];Neoadjuvant chemotherapy first, followed by chemoradiation and then surgery, in the management of locally advanced rectal cancer. J Natl Compr Canc Netw. 2014 12(4):513–519. doi: 10.6004/jnccn.2014.0056. http://www.ncbi.nlm.nih.gov/pubmed/24717570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Y-H, Lin J-Z, An X, et al. Neoadjuvant Sandwich Treatment With Oxaliplatin and Capecitabine Administered Prior to, Concurrently With, and Following Radiation Therapy in Locally Advanced Rectal Cancer: A Prospective Phase 2 Trial. Int J Radiat Oncol Biol Phys. 2014;90(5):1153–1160. doi: 10.1016/j.ijrobp.2014.07.021. doi:10.1016/j.ijrobp.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 16.Cheng DT, Mitchell T, Zehir A, et al. MSK-IMPACT: A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17(3):251–264. doi: 10.1016/j.jmoldx.2014.12.006. doi:10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyman DM, Solit DB, Arcila ME, et al. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next-generation sequencing enabling next-generation targeted therapy trials. Drug Discov Today. 2015 doi: 10.1016/j.drudis.2015.08.005. doi:10.1016/j.drudis.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duldulao MP, Lee W, Nelson RA, et al. Mutations in specific codons of the KRAS oncogene are associated with variable resistance to neoadjuvant chemoradiation therapy in patients with rectal adenocarcinoma. Ann Surg Oncol. 2013;20(7):2166–2171. doi: 10.1245/s10434-013-2910-0. doi:10.1245/s10434-013-2910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lièvre A, Bachet J-B, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. doi:10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 20.Michelassi F, Erroi F, Roncella M, Block GE. [Accessed June 8, 2015];Ras oncogene and the acquisition of metastasizing properties by rectal adenocarcinoma. Dis Colon Rectum. 1989 32(8):665–668. doi: 10.1007/BF02555770. http://www.ncbi.nlm.nih.gov/pubmed/2666052. [DOI] [PubMed] [Google Scholar]
- 21.Segelov E, Chan D, Shapiro J, et al. The role of biological therapy in metastatic colorectal cancer after first-line treatment: a meta-analysis of randomised trials. Br J Cancer. 2014;111(6):1122–1131. doi: 10.1038/bjc.2014.404. doi:10.1038/bjc.2014.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee D-W, Kim KJ, Han S-W, et al. KRAS mutation is associated with worse prognosis in stage III or high-risk stage II colon cancer patients treated with adjuvant FOLFOX. Ann Surg Oncol. 2015;22(1):187–194. doi: 10.1245/s10434-014-3826-z. doi:10.1245/s10434-014-3826-z. [DOI] [PubMed] [Google Scholar]
- 23.Yoon HH, Tougeron D, Shi Q, et al. KRAS codon 12 and 13 mutations in relation to disease-free survival in BRAF-wild-type stage III colon cancers from an adjuvant chemotherapy trial (N0147 alliance) Clin Cancer Res. 2014;20(11):3033–3043. doi: 10.1158/1078-0432.CCR-13-3140. doi:10.1158/1078-0432.CCR-13-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clancy C, Burke JP, Coffey JC. KRAS mutation does not predict the efficacy of neo-adjuvant chemoradiotherapy in rectal cancer: a systematic review and meta-analysis. Surg Oncol. 2013;22(2):105–111. doi: 10.1016/j.suronc.2013.02.001. doi:10.1016/j.suronc.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Gaedcke J, Grade M, Jung K, et al. KRAS and BRAF mutations in patients with rectal cancer treated with preoperative chemoradiotherapy. Radiother Oncol. 2010;94(1):76–81. doi: 10.1016/j.radonc.2009.10.001. doi:10.1016/j.radonc.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.