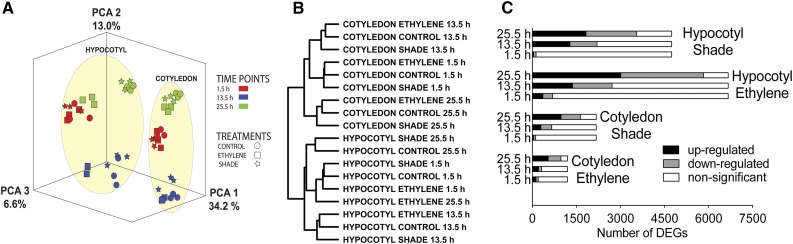
Figure 2.
Overall description of microarray data. A, Principal component analysis (PCA) and hierarchical clustering were used to describe the structure in the microarray data. Expression intensities for all genes on the array for all 54 hypocotyl and cotyledon samples (three time points, three treatments, and three replicates) were projected onto the first three principal components. B, Hierarchical clustering was used to group 18 main samples (according to the mean expression intensity of three replicates for each main sample) into a dendrogram. C, Distribution of DEGs in hypocotyl and cotyledon samples at three time points in response to ethylene and shade. DEGs obtained for each time point were plotted separately as up-regulated, down-regulated (adjusted P ≤ 0.01 and log2FC greater than or less than 0), or nonsignificant (adjusted P > 0.01). Bar length denotes total DEGs obtained after combining DEGs from all three time points.