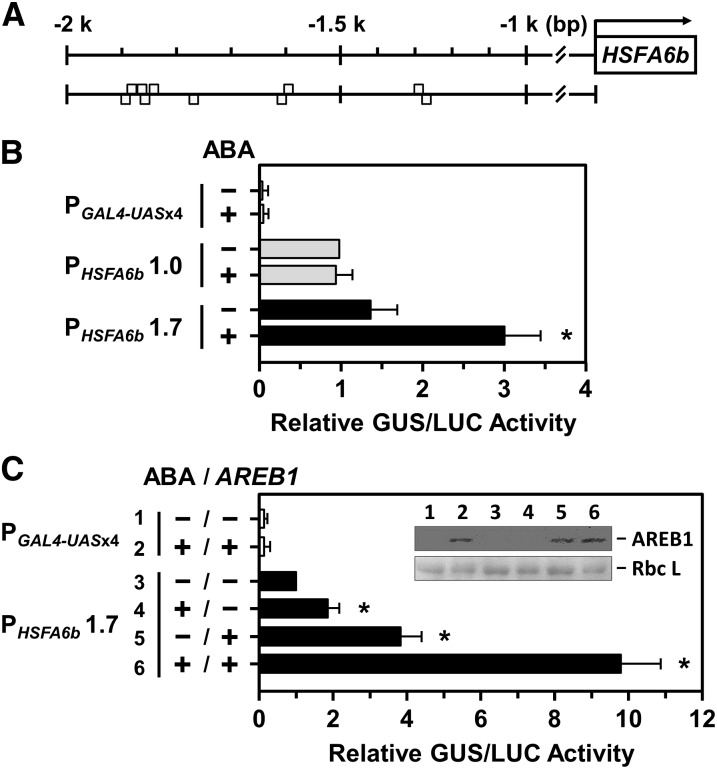
Figure 4.
The transcriptional activation of the HSFA6b promoter in response to ABA and AREB1 treatments were analyzed in mesophyll protoplasts. A, Gray boxes are predicted ABRE motifs (T/CACGTGGC) upstream of the HSFA6b promoter region at −1257 (reverse strand [−]); −1299 (forward strand [+]); −1602 (−); −1605 (+); −1776 (−); −1846 (−); −1849 (+); −1864 (+); −1869 (−); and −1872 bp (+). B and C, The transcriptional activation assay of the HSFA6b promoter. PGAL4-UASx4::GUS was a control, and 1- or 1.7-kb lengths of the HSFA6b promoter region, PHSFA6b1.0 or 1.7, respectively, were fused with GUS gene and used in the corresponding transient reporter assays. Transfected protoplasts were then treated with (+) or without (−) 10 μm ABA for 12 h and with or without effector AREB1-3XFLAG coexpression, as indicated. The AREB1-3XFLAG fusion protein, under the control of the CaMV 35S promoter, was detected by immunoblotting (α-FLAG) and the Rubisco large subunit (Rbc L) shown for equal loading (inset). The amount of relative GUS activity was normalized by luciferase (LUC) luminescence. The fold expression was normalized relative to the amount of PHSFA6b 1.0 without ABA (B), or PHSFA6b 1.7 without ABA and ARER1 (C). Data are means ± sd of three biological replicates. *Significant at P < 0.05.