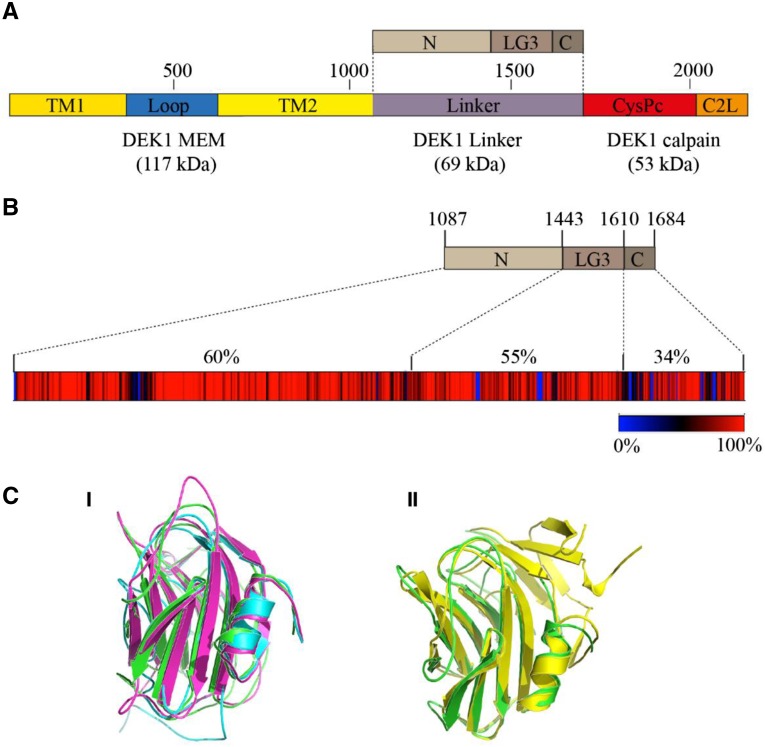
Figure 1.
P. patens DEK1 protein domain structure and DEK1 Linker sequence conservation. A, Schematic representation of DEK1 domain structure. DEK1 MEM is composed of two parts, with multiple TM segments disrupted by the Loop. The DEK1 Linker is divided into the three subsegments, N, LG3, and C. DEK1 calpain consists of CysPc, the catalytic domain, and C2L. Amino acid numbering is given above the DEK1 outline. B, Heat map showing the degree of DEK1 Linker sequence conservation among 73 land plant species. Percentage sequence identities are indicated in different colors, from blue (0%; insertions/deletions) to red (100%; absolute conserved positions). Amino acid numbering for the different subsegments of the P. patens DEK1 Linker is given above the top part. Percentages above the heat map give the sequence identities between the Arabidopsis and P. patens DEK1 Linker subsegments. C, Predicted structure of the P. patens DEK1 LG3 domain. I, Structural alignment of the predicted I-TASSER (blue), RaptorX (magnenta), and Phyre2 (green) structures showing that the LG3 domain adopts a β-sandwich fold and that individual servers predicted highly similar structures. II, Structural alignment between the Phyre2 model and the Protein Data Base (PDB) structure 3FLP (yellow) of native heptameric serum amyloid P component (SAP)-like pentraxin from Limulus polyphemus.