A glycosylphosphatidylinositol modification is a necessary and sufficient signal for enriched protein accumulation in a plasma membrane domain associated with plasmodesmata.
Abstract
Plasmodesmata (Pd) are membranous channels that serve as a major conduit for cell-to-cell communication in plants. The Pd-associated β-1,3-glucanase (BG_pap) and CALLOSE BINDING PROTEIN1 (PDCB1) were identified as key regulators of Pd conductivity. Both are predicted glycosylphosphatidylinositol-anchored proteins (GPI-APs) carrying a conserved GPI modification signal. However, the subcellular targeting mechanism of these proteins is unknown, particularly in the context of other GPI-APs not associated with Pd. Here, we conducted a comparative analysis of the subcellular targeting of the two Pd-resident and two unrelated non-Pd GPI-APs in Arabidopsis (Arabidopsis thaliana). We show that GPI modification is necessary and sufficient for delivering both BG_pap and PDCB1 to Pd. Moreover, the GPI modification signal from both Pd- and non-Pd GPI-APs is able to target a reporter protein to Pd, likely to plasma membrane microdomains enriched at Pd. As such, the GPI modification serves as a primary Pd sorting signal in plant cells. Interestingly, the ectodomain, a region that carries the functional domain in GPI-APs, in Pd-resident proteins further enhances Pd accumulation. However, in non-Pd GPI-APs, the ectodomain overrides the Pd targeting function of the GPI signal and determines a specific GPI-dependent non-Pd localization of these proteins at the plasma membrane and cell wall. Domain-swap analysis showed that the non-Pd localization is also dominant over the Pd-enhancing function mediated by a Pd ectodomain. In conclusion, our results indicate that segregation between Pd- and non-Pd GPI-APs occurs prior to Pd targeting, providing, to our knowledge, the first evidence of the mechanism of GPI-AP sorting in plants.
Plant cells are interconnected with cross-wall membranous channels called plasmodesmata (Pd). Recent studies have shown that the region of the plasma membrane (PM) lining the Pd channel is a specialized membrane microdomain whose lipid and protein composition differs from the rest of the PM (Tilsner et al., 2011, 2016; Bayer et al., 2014; González-Solís et al., 2014; Grison et al., 2015). In a similar manner, the cell wall domain surrounding the Pd channel is specialized and, unlike the rest of the cell wall, is devoid of cellulose, rich in pectin, and contains callose (an insoluble β-1,3-glucan; Zavaliev et al., 2011; Knox and Benitez-Alfonso, 2014). In response to physiological signals, callose can be transiently deposited and degraded at Pd, which provides a mechanism for controlling the Pd aperture in diverse developmental and stress-related processes (Zavaliev et al., 2011). Control of Pd functioning is mediated by proteins that are specifically targeted to Pd. Plasmodesmal proteins localized to the PM domain of Pd can be integral transmembrane proteins, such as Pd-localized proteins (Thomas et al., 2008), the receptor kinase ARABIDOPSIS CRINKLY4 (Stahl et al., 2013), and callose synthases (Vatén et al., 2011). Alternatively, Pd proteins can associate with the membrane through a lipid modification like myristoylation (e.g. remorins; Raffaele et al., 2009) or be attached by a glycosylphosphatidylinositol (GPI) anchor (e.g. Pd-associated β-1,3-glucanases [BG_pap]; Levy et al., 2007; Rinne et al., 2011; Benitez-Alfonso et al., 2013), Pd-associated callose-binding proteins (PDCBs; Simpson et al., 2009), and LYSIN MOTIF DOMAIN-CONTAINING PROTEIN2 (LYM2; Faulkner et al., 2013).
Among the known Pd proteins involved in Pd-specific callose degradation is BG_pap, a cell wall enzyme carrying a glycosyl hydrolase family 17 (GH17) module as its functional domain (Levy et al., 2007). Another group of proteins controlling callose dynamics at Pd are PDCBs that harbor a callose-binding domain termed carbohydrate-binding module 43 (CBM43), implicated in stabilizing callose at Pd (Simpson et al., 2009). Some β-1,3-glucanases may combine the two callose-modifying activities by harboring both GH17 and CBM43 functional domains, and several such proteins were shown to localize to Pd (Rinne et al., 2011; Benitez-Alfonso et al., 2013; Gaudioso-Pedraza and Benitez-Alfonso, 2014). A distinct feature of BG_pap and PDCBs is that both are predicted glycosylphosphatidylinositol-anchored proteins (GPI-APs). The GPI anchor is a form of posttranslational modification common to many cell surface proteins in all eukaryotes. GPI-APs are covalently attached to the outer leaflet of the PM through the GPI anchor. The basic structure of the anchor consists of ethanolamine phosphate, followed by a glycan chain of three Man residues and glucosamine, followed by phosphatidylinositol lipid moiety (EtNP-6Manα1-2Manα1-6Manα1-4GlcNα1-6myoinositol-1-P-lipid; Ferguson et al., 2009). All predicted GPI-APs carry an N-terminal secretion signal peptide (SP) similar to other secreted proteins. Distinctly, GPI-APs also carry a structurally conserved 25- to 30-residue C-terminal GPI attachment signal, which typically begins with a small amino acid (e.g. Ala, Asn, Asp, Cys, Gly, or Ser) termed omega, followed by a spacer region of five to 10 polar residues, and ending with a transmembrane segment of 15 to 20 hydrophobic residues (Ferguson et al., 2009). The entire region between the N-terminal and the C-terminal signals of a GPI-AP is termed the ectodomain and carries the protein’s functional domain(s). The GPI modification process takes place in the lumenal face of the endoplasmic reticulum (ER) in a cotranslational manner. Upon translocation into the ER, a GPI-AP is stabilized in the ER membrane by its C-terminal signal, which is concurrently cleaved after the omega amino acid, and a preassembled GPI anchor is covalently attached to the C terminus of the omega amino acid. After attachment to a protein, the GPI anchor undergoes a series of modifications (remodeling), both at the glucan chain and at the lipid moiety. Such remodeling is crucial for the sorting of GPI-APs in the secretory pathway and the subsequent lateral heterogeneity at the PM (Kinoshita, 2015). In particular, the addition of saturated fatty acid chains to the lipid moiety of the anchor leads to the enriched accumulation of GPI-APs in the PM microdomains, also termed lipid rafts (Muñiz and Zurzolo, 2014). In Arabidopsis (Arabidopsis thaliana), GPI modification has been predicted for 210 proteins of diverse functions at the PM or the cell wall or both (Borner et al., 2002). Despite extensive research on the GPI modification pathway and the function of GPI-APs in mammalian and yeast cells, such knowledge in plant systems is scarce. In particular, despite an emerging role of GPI-APs in the regulation of the cell wall domain of Pd, their subcellular targeting and compartmentalization mechanism have not been studied. In addition, it is not known how the targeting mechanism of Pd-resident GPI-APs is different from that of other classes of GPI-APs, which are not localized to Pd.
In this study, we investigated the subcellular targeting mechanism of Pd-associated callose-modifying GPI-APs, BG_pap and PDCB1, and compared it with that of two unrelated non-Pd GPI-APs, ARABINOGALACTAN PROTEIN4 (AGP4) and LIPID TRANSFER PROTEIN1 (LTPG1). Using sequential fluorescent labeling of protein domains, we found that the C-terminal GPI modification signal present in both Pd- and non-Pd GPI-APs can function as a primary signal in targeting proteins to the Pd-enriched PM domain. Moreover, we show that while the GPI signal is sufficient for Pd targeting, the ectodomains in BG_pap and PDCB1 further enhance their accumulation at Pd. In contrast, the ectodomains in non-Pd GPI-APs mediate exclusion of the proteins from the Pd-enriched targeting pathway. The Pd exclusion effect was found to be dominant over the Pd-targeting function of the GPI signal and the Pd-enhancing function of the Pd ectodomain, and it possibly occurs prior to PM localization. Our findings thus uncover a novel Pd-targeting signal and provide, to our knowledge, the first evidence of the cellular mechanism that regulates the sorting of GPI-APs in plants.
RESULTS
GPI Modification Is Required for Subcellular Targeting of Callose-Modifying BG_pap and PDCB1
To identify factors regulating the subcellular targeting of Pd-associated GPI-APs, we chose two previously reported callose-modifying proteins, BG_pap (Levy et al., 2007) and PDCB1 (Simpson et al., 2009), for the analysis. For comparison with GPI-APs that do not have a known Pd localization (non-Pd GPI-APs) we included two unrelated previously reported GPI-APs, the cell surface protein AGP4 (Sherrier et al., 1999) and the lipid transfer protein LTPG1 (Debono et al., 2009; Ambrose et al., 2013). All four GPI-APs carry a predicted C-terminal GPI modification signal with Ser as an omega amino acid (Fig. 1A). The proteins were tagged with monomeric citrine (mCitrine; Zacharias et al., 2002), which is more suitable for extracellular proteins in plant cells due to improved fluorescent properties at acidic pH. The mCitrine tag was placed at the N terminus, after the SP (Fig. 1B). To achieve uniformity in secretion and facilitate comparisons between different GPI-APs and their segments, we used the SP sequence from the Arabidopsis chitinase gene, which is commonly used to drive the secretion of tagged proteins in plant cells (Haseloff et al., 1997; Frigerio et al., 2001; Gidda et al., 2009). The localization of each fusion protein was tested using transient expression in Nicotiana benthamiana and confirmed in transgenic Arabidopsis. Localization of the full-length fusions constructed with the SP sequence from the Arabidopsis chitinase gene was similar to the previously published localizations of these proteins. The sizes of the fusion proteins were confirmed by western blotting (Supplemental Fig. S1). To verify GPI modification, we used mannosamine, an amino sugar that inhibits the biosynthesis of the GPI anchor by incorporating into the second Man position of the glycan portion of the GPI anchor (Lisanti et al., 1991; Smith, 2009). By depleting cells of the mature GPI substrate, mannosamine leads to the accumulation of GPI-APs in compartments along the secretion pathway, either in the ER or the Golgi or both (Lisanti et al., 1991; Pan et al., 1992; Sevlever and Rosenberry, 1993; De Caroli et al., 2011).
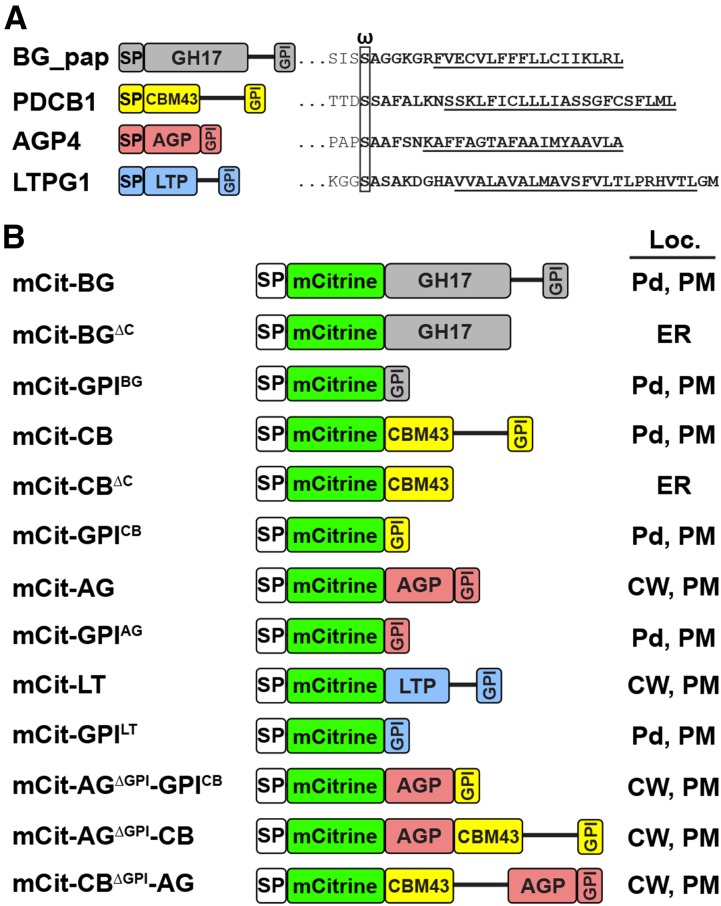
Figure 1.
List of GPI-anchored proteins characterized in this study. A, Left, the GPI-anchored proteins BG_pap, PDCB1, AGP4, and LTPG1. Right, details of the C-terminal region, including the predicted GPI modification signal (boldface). The omega amino acid is boxed and the transmembrane segment is underlined. B, Names and diagrams of SP-mCitrine fusions of the proteins listed in A. The SP is not included in the fusion names, and the protein names are abbreviated: BG, BG_pap; CB, PDCB1; AG, AGP4; LT, LTPG1. The determined subcellular localization (Loc.) for each fusion is shown at right: CW, cell wall.
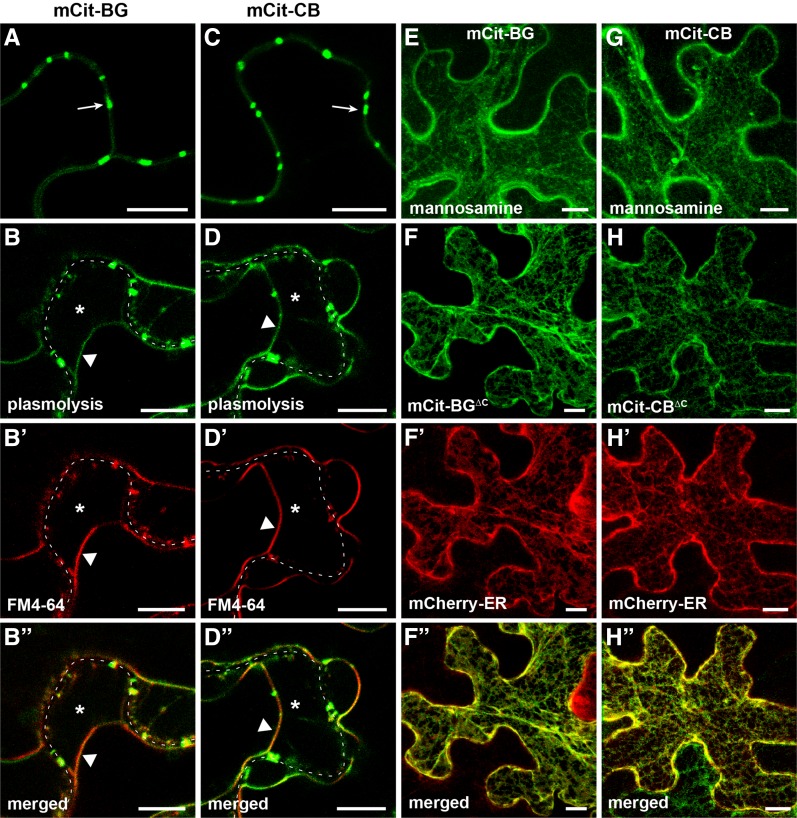
Full-length BG_pap (hereafter a protein sequence without the native SP) fused to SP-mCitrine (mCit-BG) showed enriched accumulation at peripheral puncta (Fig. 2A), consistent with its previously reported localization (Levy et al., 2007). The mCit-BG also showed accumulation at the PM, as evident from plasmolysis and staining with FM4-64 (Fig. 2, B–B′′). The appearance of the PM localization is possibly a result of overexpression and is expected, since BG_pap is a predicted GPI-AP. Like BG_pap, full-length PDCB1 fused to SP-mCitrine (mCit-CB) showed enriched accumulation at peripheral puncta (Fig. 2C), consistent with its previously reported localization (Simpson et al., 2009). In plasmolyzed cells, mCit-CB showed a PM fraction as well (Fig. 2, D–D′′). Application of 10 mm mannosamine significantly reduced the targeting of both mCit-BG and mCit-CB and led to intracellular accumulation in the ER-like and vesicle-like structures (Fig. 2, E and G). Next, we sought to determine whether the callose-modifying modules in BG_pap and PDCB1, GH17BG_pap and CBM43PDCB1, respectively, are able to target to Pd as soluble proteins. To this end, the C-terminal part including the GPI signal was deleted in the full-length fusion proteins (Fig. 1B). The resulting mCit-BGΔC and mCit-CBΔC fusions did not show localization at peripheral puncta but had predominant ER accumulation, as evident from colocalization with mCherry-ER (Fig. 2, F–F′′ and H–H′′). Together, these results demonstrate that GPI modification is required for the subcellular targeting of BG_pap and PDCB1 and that the functional domains in both proteins are not delivered to peripheral puncta in the absence of a GPI signal.
Figure 2.
GPI modification is required for the targeting of BG_pap and PDCB1. Localization is shown for BG_pap and PDCB1 in transient expression in N. benthamiana. A to B′′, Localization of SP-mCitrine fused to full-length BG_pap (mCit-BG). C to D′′, Localization of SP-mCitrine fused to full-length PDCB1 (mCit-CB). E and G, Localization of mCit-BG (E) and mCit-CB (G) in the presence of the GPI biosynthesis inhibitor mannosamine. F to F′′, Localization of SP-mCitrine fused to BG_pap in which the C-terminal part is deleted (mCit-BGΔC). H to H′′, Localization of SP-mCitrine fused to PDCB1 in which the C-terminal part is deleted (mCit-CBΔC). B to B′′ and D to D′′ show localization in plasmolyzed cells stained with FM4-64. F to F′′ and H to H′′ show colocalization with the ER marker mCherry-ER. Arrows in A and C indicate peripheral puncta. In plasmolyzed cells, the dashed white line marks the cell wall, asterisks indicate the apoplastic space formed by the receding protoplast, and triangles indicate the position of the PM. Images in E to H′′ are Z projections of several confocal slices. Bars = 10 µm.
The C-Terminal GPI Modification Signal from Both Pd- and Non-Pd GPI-Anchored Proteins Mediates Pd Targeting
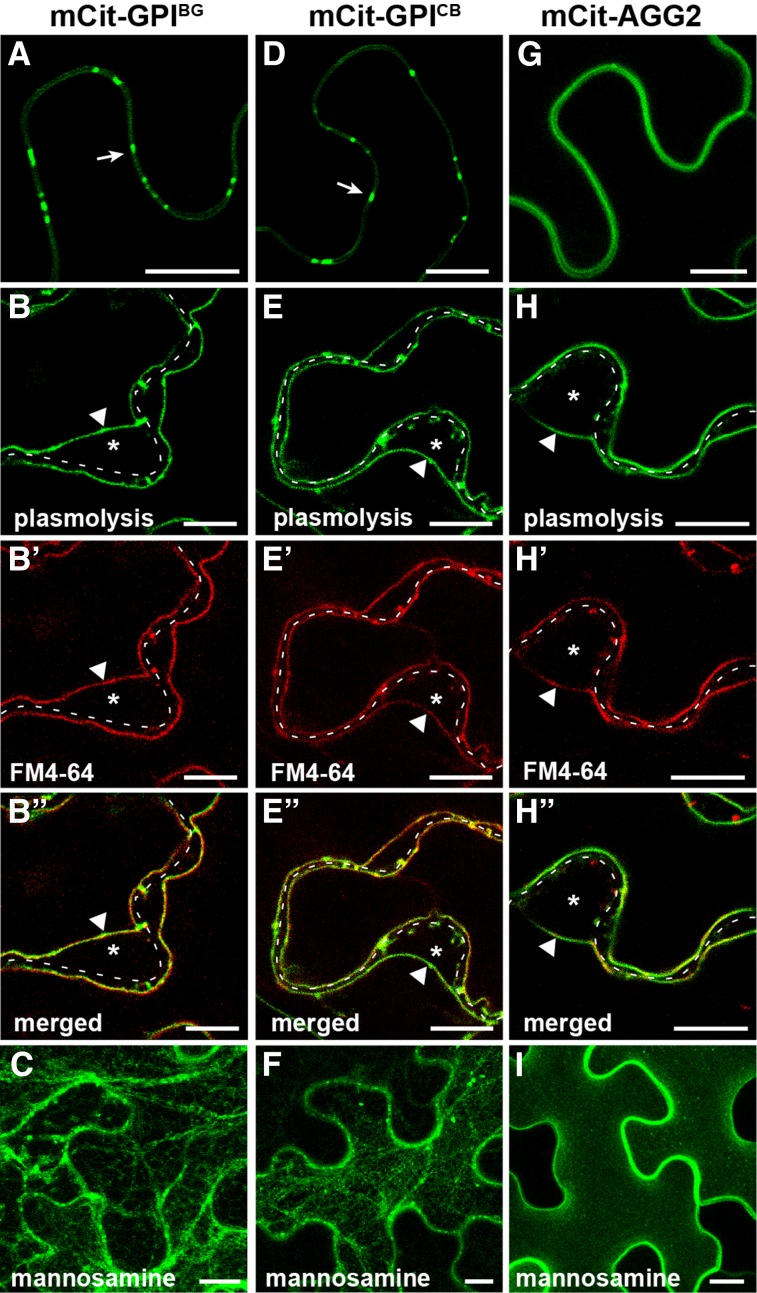
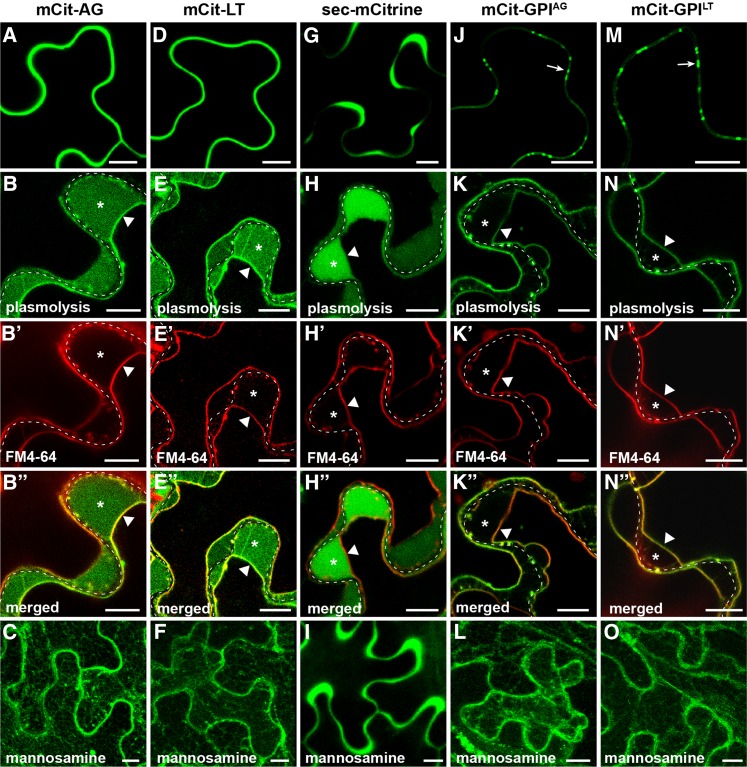
To further understand the role of GPI modification in Pd targeting of BG_pap and PDCB1, the entire ectodomain in both proteins was deleted and SP-mCitrine was attached to a predicted omega amino acid of the GPI signal (Fig. 1B). Interestingly, both mCit-GPIBG and mCit-GPICB showed enriched accumulation at peripheral puncta in a pattern similar to that of the full-length proteins (Fig. 3, A and D), suggesting a possible Pd localization. Like the full-length proteins, mCit-GPIBG and mCit-GPICB also showed accumulation at non-Pd PM (Fig. 3, B–B′′ and E–E′′). Mannosamine inhibited the export of both fusions and led to ER-like and vesicle-like accumulation (Fig. 3, C and F), confirming the GPI-dependent targeting. As a proof of concept of mannosamine action, further deletion of the region containing the omega site and attachment of SP-mCitrine to the transmembrane segment of the GPI signal fully blocked protein targeting and resulted in the ER accumulation (Supplemental Fig. S2). To rule out the possibility that a Pd-like enrichment of GPI-anchored mCitrine is an artifact of a stress response at the PM associated with agroinfiltration, we tested the localization of a non-GPI PM protein in our expression system. As a control, we chose previously reported ARABIDOPSIS HETEROTRIMERIC G-PROTEIN γ-SUBUNIT2 (AGG2) that is a prenylated PM protein (Zeng et al., 2007). Expression of mCitrine fused to AGG2 (mCit-AGG2) resulted in a uniform PM accumulation with no enrichment at discrete regions (Fig. 3G), indicating that the localization of GPI-anchored mCitrine is not a result of unspecific changes in membrane architecture. The PM localization of mCit-AGG2 was confirmed by plasmolysis and FM4-64 staining (Fig. 3, H–H′′). Unlike GPI-anchored fusions, the targeting of mCit-AGG2 was not inhibited by mannosamine (Fig. 3I), providing evidence that the effect of mannosamine on GPI-APs is not a result of nonspecific response to the drug.
Figure 3.
GPI modification signal mediates enriched accumulation at the peripheral puncta. Transient expression is shown for GPI-anchored mCitrine in N. benthamiana. A to C, Localization of SP-mCitrine fused to the GPI signal of BG_pap (mCit-GPIBG). D to F, Localization of SP-mCitrine fused to the GPI signal of PDCB1 (mCit-GPICB). G to I, Localization of mCitrine fused to the PM marker AGG2 (mCit-AGG2). A, D, and G, show localization with no treatment; B to B′′, E to E′′, and H to H′′ show localization in plasmolyzed cells stained with FM4-64. C, F, and I, show localization in the presence of the GPI biosynthesis inhibitor mannosamine. Arrows in A and D indicate peripheral puncta. In plasmolyzed cells, the dashed white line marks the cell wall, asterisks indicate the apoplastic space formed by the receding protoplast, and triangles indicate the position of the PM. Images in C, F, and I are Z projections of several confocal slices. Bars = 10 µm.
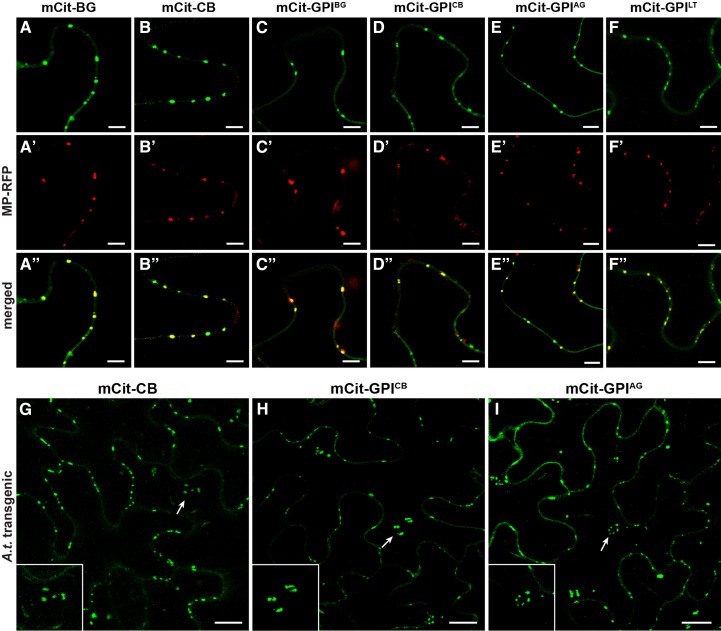
To find out whether GPI-anchored mCitrine is indeed localized to Pd, we tested its colocalization with the movement protein of Tobacco mosaic virus fused to red fluorescent protein (MP-RFP), which is commonly used as a Pd marker (Boutant et al., 2010). As established Pd-associated proteins, both mCit-BG and mCit-CB fusions, showed full colocalization with MP-RFP at Pd (Fig. 4, A–A′′ and B–B′′). Importantly, the minimal fusions mCit-GPIBG and mCit-GPICB also fully colocalized with MP-RFP (Fig. 4, C–C′′ and D–D′′), indicating accumulation at Pd sites. The observed ability of GPI signals from BG_pap and PDCB1 to mediate Pd localization of the reporter protein raised the question of whether such function is true for GPI signals from non-Pd GPI-APs. To this end, we tested GPI signals from the two non-Pd GPI-APs, AGP4 and LTPG1, for their ability to mediate Pd targeting of mCitrine. As before, SP-mCitrine was fused to the omega amino acid of the predicted GPI signal in both AGP4 and LTPG1 (Fig. 1B). Similar to GPI signals from Pd-associated GPI-APs, both mCit-GPIAG and mCit-GPILT showed accumulation at peripheral puncta that fully colocalized with MP-RFP (Fig. 4, E–E′′ and F–F′′), indicating Pd localization. The Pd targeting of GPI-anchored mCitrine also was observed in transgenic Arabidopsis (Fig. 4, H and I). Similar to full-length fusion mCit-CB (Fig. 4G), both mCit-GPICB and mCit-GPIAG showed accumulation at peripheral puncta characteristic of Pd (Fig. 4, H and I). Moreover, unlike in transient expression, localization in transgenic plants could be detected at periclinal Pd clusters connecting epidermis with mesophyll (Fig. 4, G–I, insets). The Pd localization in transgenic plants also was confirmed for mCit-BG, mCit-GPIBG, and mCit-GPILT (Supplemental Fig. S3). Together, these results reveal that the C-terminal GPI signal from Pd- as well as non-Pd GPI-APs can mediate the localization of a reporter protein to Pd, indicating that GPI modification is a sufficient signal for Pd targeting.
Figure 4.
GPI-anchored mCitrine is localized to Pd. A to F′′, Colocalization of full-length GPI-APs and GPI-anchored mCitrine with MP-RFP in transient expression in N. benthamiana: mCit-BG (A–A′′), mCit-CB (B–B′′), mCit-GPIBG (C–C′′), mCit-GPICB (D–D′′), mCit-GPIAG (E–E′′), and mCit-GPILT (F–F′′). G to I, Localization of mCit-CB (G), mCit-GPICB (H), and mCit-GPIAG (I) in transgenic Arabidopsis. Images in G to I show Z projections of several confocal slices across the entire epidermal cell, including both anticlinal and periclinal cell surfaces. Arrows in G to I indicate the positions of the periclinal Pd cluster (enlarged in the insets). Bars = 5 µm (A–F′′) and 10 µm (G–I).
Ectodomain in Pd-Associated GPI-Anchored Proteins Enhances Pd Accumulation
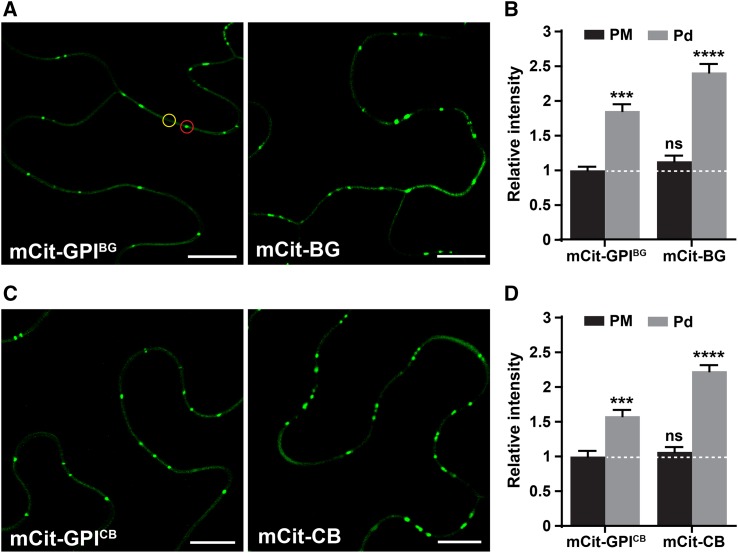
To determine the effect of an ectodomain on the Pd targeting mediated by a GPI signal, we compared the level of Pd and PM accumulation of the full-length BG_pap and PDCB1 fusions with that of GPI-anchored mCitrine (Fig. 5). To achieve uniform expression for each fusion protein and to avoid variations in tissue state, the analysis was performed using a transient expression assay in N. benthamiana half-leaves. The full-length fusion was expressed in one half of a leaf and the corresponding GPI-anchored mCitrine in the other half: mCit-BG and mCit-GPIBG (Fig. 5A) or mCit-CB and mCit-GPICB (Fig. 5C). For each fusion protein, the fluorescence intensity was measured from at least 15 images at two distinct regions: at the non-Pd region (designated as PM) and at the Pd foci (designated as Pd; Fig. 5A, circles). The obtained PM and Pd intensities for each fusion protein are shown as the ratio of the PM intensity measured for the corresponding GPI-anchored mCitrine (Fig. 5, B and D). Relative to PM accumulation, both GPI-anchored mCitrine and full-length fusions showed significant enrichment at Pd (Fig. 5, B and D). Notably, the Pd enrichment of the full-length fusions was significantly higher than that of GPI-anchored mCitrine (Fig. 5, B and D). In contrast, the PM fraction in the full-length fusions was not significantly different from that of the corresponding GPI-anchored mCitrine (Fig. 5, B and D). These results indicate that ectodomains in BG_pap and PDCB1 enhance protein accumulation above the level mediated by their GPI signals, specifically at the Pd but not at the PM. Such increased Pd accumulation could result from the interaction of the ectodomains in BG_pap and PDCB1 with a Pd component, such as callose. A supportive piece of evidence for such an interaction could be observed in cells treated with the stress hormone salicylic acid (SA; Supplemental Fig. S4). It has been shown that GPI-APs are susceptible to cleavage and release from the PM in response to stress (Ferguson et al., 2009; Simpson et al., 2009). To achieve such an effect, we treated leaves expressing mCit-BG, mCit-GPIBG, and the control PM protein mCit-AGG2 with 5 mm SA and followed protein localization. Unlike control PM protein, SA treatment triggered release of the full-length mCit-BG from the PM to the apoplast but had little effect on the Pd fraction (Supplemental Fig. S4). In contrast, a complete release of mCit-GPIBG from the PM correlated with a complete loss of the Pd fraction (Supplemental Fig. S4). Although SA-mediated stress signaling may have multiple effects on the posttranslational modification and targeting of proteins, these observations nevertheless support the possibility of ectodomain interaction with a Pd component and subsequent enhanced accumulation of the full-length BG_pap and PDCB1 at Pd.
Figure 5.
Ectodomain in BG_pap and PDCB1 enhances Pd accumulation. Pd and PM accumulation levels are shown for the full-length fusions compared with those of GPI-anchored mCitrine in transient expression in N. benthamiana. A and C, Representative confocal images of cells expressing mCit-GPIBG and mCit-BG (A) or mCit-GPICB and mCit-CB (C). B and D, Relative fluorescence intensities quantified for each fusion protein at two distinct regions as indicated in A, PM (yellow circle) and Pd (red circle). Values represent mean fluorescence intensity at PM or Pd relative to mean fluorescence intensity at PM measured for mCit-GPIBG (B) and mCit-GPICB (D). The white dashed lines in B and D represent the reference line corresponding to 1. Error bars represent se. Lowercase letters indicate statistically significant differences determined by nonparametric Kruskal-Wallis one-way ANOVA test for multiple groups (***P < 0.001; ****P < 0.0001; ns=not significant). The experiment was performed two times with similar results. Bars = 10 μm.
Ectodomain in Non-Pd GPI-Anchored Proteins Overrides the Pd-Targeting Function of the GPI Signal and the Pd-Enhancing Function of the Pd Ectodomain
As we showed in a previous section, GPI signals of the two non-Pd GPI-APs, AGP4 and LTPG1, when expressed in the absence of their respective ectodomains mediate Pd targeting of mCitrine (Fig. 4, E and F). Next, we sought to determine how the ectodomains in AGP4 and LTPG1 affect the Pd-targeting function of their GPI signals. The ectodomain in AGP4 is an 85-amino acid-long polypeptide (AGP motif; Fig. 1A) that serves as a backbone for extensive O-linked glycosylations at multiple Pro residues (Sherrier et al., 1999; Ellis et al., 2010). The ectodomain in LTPG1 is a 127-amino acid-long polypeptide carrying a lipid transfer protein as a functional domain (Fig. 1A) that has been shown to have lipid-binding activity (Debono et al., 2009). Fusion of SP-mCitrine to full-length AGP4 (mCit-AG) resulted in a uniform accumulation at the cell periphery (Fig. 6A). Plasmolysis and FM4-64 staining revealed a PM localization along with a soluble apoplastic fraction in mCit-AG (Fig. 6, B–B′′), indicating that the protein is both PM and cell wall localized. Such localization is consistent with a previously reported localization of Arabidopsis AGPs fused to GFP (Zhang et al., 2011a). Mannosamine treatment blocked the export of mCit-AG (Fig. 6C), indicating the GPI-dependent targeting. Fusion of SP-mCitrine to full-length LTPG1 (mCit-LT) showed a similar uniform peripheral localization pattern (Fig. 6D) and appeared both at the PM and the cell wall (Fig. 6, E–E′′), consistent with previously reported localizations of this GPI-AP (Debono et al., 2009; Ambrose et al., 2013). Mannosamine treatment blocked the export of mCit-LT (Fig. 6F), indicating GPI-dependent targeting. To compare the accumulation pattern of AGP4 and LTPG1 with that of a fully soluble apoplastic protein, we tested the localization of secreted soluble mCitrine (sec-mCitrine) in our expression system (Fig. 6, G–I). Unlike, mCit-AG and mCit-LT, sec-mCitrine showed a highly nonuniform peripheral localization pattern (Fig. 6G), which corresponded to a fully apoplastic accumulation in plasmolyzed cells with no detectable PM fraction (Fig. 6, H–H′′). Mannosamine did not affect the targeting of sec-mCitrine (Fig. 6I), indicating that the drug does not detectibly affect the export of a soluble secreted protein. These results indicate that the cell wall fraction of mCit-AG and mCit-LT most likely originates from the GPI-anchored PM fraction rather than from secretion of a soluble form of the proteins and/or that of free mCitrine. Unlike the full-length AGP4 and LTPG1, their GPI signals fused to SP-mCitrine, mCit-GPIAG and mCit-GPILT, respectively, led to enriched Pd accumulation of the reporter protein (Fig. 6, J and M). Similar to GPI signals from the Pd proteins, both mCit-GPIAG and mCit-GPILT localized at the PM with no detectable cell wall fraction (Fig. 6, K–K′′ and N–N′′) and were inhibited by mannosamine (Fig. 6, L and O). In transgenic plants, both full-length AGP4 and LTPG1, as well as their GPI signals, showed similar localizations as in the transient expression (Fig. 4; Supplemental Fig. S3). Together, these results show that ectodomains in the two non-Pd GPI-APs, AGP4 and LTPG1, override the Pd-targeting function of their GPI signals and determine a specific GPI-dependent non-Pd localization of these proteins.
Figure 6.
Non-Pd ectodomain overrides the Pd-targeting function of the GPI signal. Subcellular localization is shown for AGP4 and LTPG1 in transient expression in N. benthamiana. A to C, Localization of SP-mCitrine fused to full-length AGP4 (mCit-AG). D to F, Localization of SP-mCitrine fused to full-length LTPG1 (mCit-LT). G to I, Localization of sec-mCitrine. J to L, Localization of SP-mCitrine fused to GPI signal of AGP4 (mCit-GPIAG). M to O, Localization of SP-mCitrine fused to GPI signal of LTPG1 (mCit-GPILT). A, D, G, J, and M show localization with no treatment; B to B′′, E to E′′, H to H′′, K to K′′, and N to N′′ show localization in plasmolyzed cells stained with FM4-64; C, F, I, L, and O show localization in the presence of the GPI biosynthesis inhibitor mannosamine. In plasmolyzed cells, the dashed white line marks the cell wall, asterisks indicate the apoplastic space formed by the receding protoplast, and triangles indicate the position of PM. Arrows in J and M indicate the positions of Pd foci. Images in C, F, I, L, and O are Z projections of several confocal slices. Bars = 10 µm.
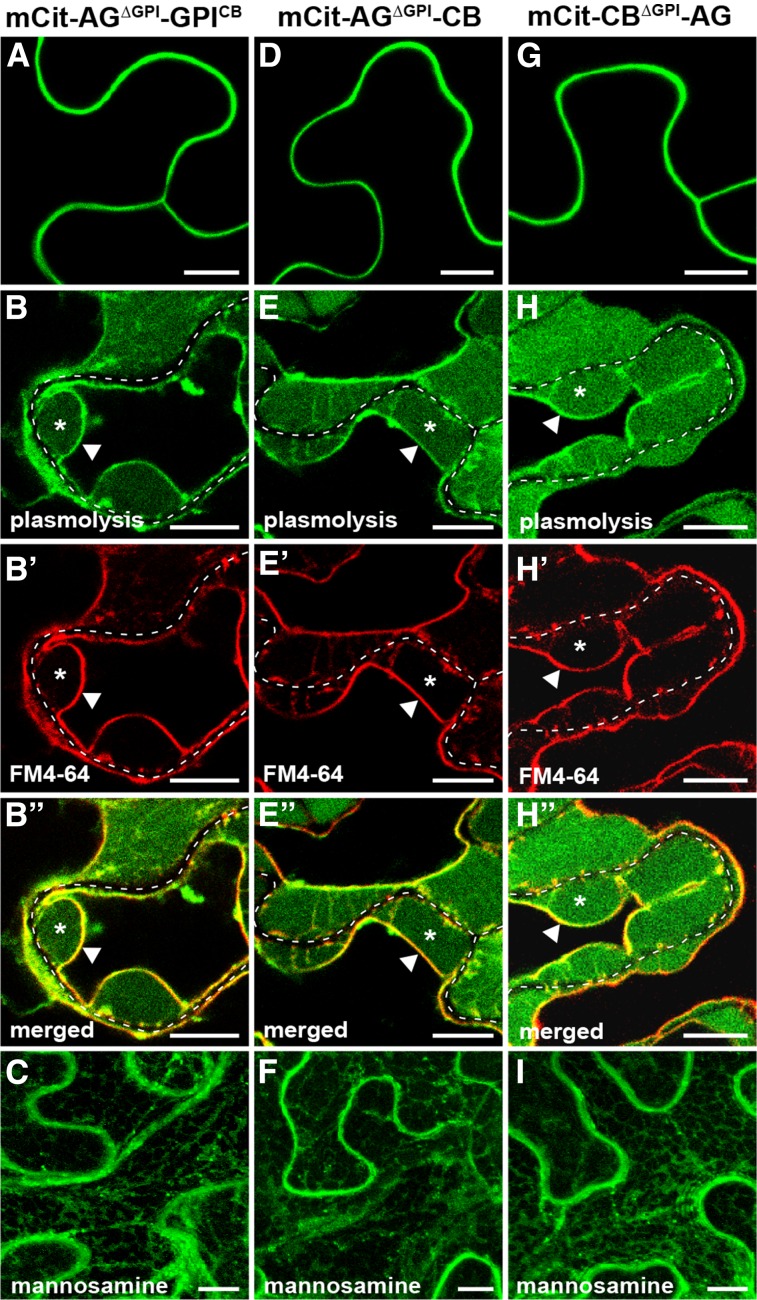
To test whether a non-Pd ectodomain can similarly override the Pd targeting of a Pd-associated GPI-AP, we created chimeric fusions between AGP4 and PDCB1 (Fig. 1B). We chose AGP4 and PDCB1 due to their smaller sizes compared with LTPG1 and BG_pap, respectively (Fig. 1A). First, the GPI signal in the mCit-AG fusion was replaced with GPI signal of PDCB1, creating mCit-AGΔGPI-GPICB (Fig. 1B). This chimeric fusion showed a non-Pd localization pattern like that of mCit-AG (Fig. 7A), had similar PM and cell wall fractions (Fig. 7, B–B′′), and was inhibited by mannosamine (Fig. 7C), confirming GPI-dependent targeting. These results indicate that the Pd exclusion effect of a non-Pd ectodomain is equally dominant over GPI signal taken from a Pd-resident protein. Next, we sought to determine how the Pd exclusion mediated by a non-Pd ectodomain is affected in the presence of a Pd ectodomain. To this end, the GPI signal in mCit-AG was replaced with the ectodomain and GPI signal of PDCB1 to create mCit-AGΔGPI-CB (Fig. 1B). In the reciprocal fusion, GPI signal in mCit-CB was replaced with ectodomain and GPI signal of AGP4 to create mCit-CBΔGPI-AG (Fig. 1B). Interestingly, both chimeric fusions showed a localization pattern similar to that of mCit-AG and had no detectable Pd enrichment (Fig. 7, D and G). Both chimeras had accumulation at the PM and cell wall (Fig. 7, E–E′′ and H–H′′) and were inhibited by mannosamine (Fig. 7, F and I), confirming GPI-dependent targeting. Together, these results show that the Pd exclusion effect of a non-Pd ectodomain is dominant over the Pd-targeting function of a GPI signal as well as the Pd-enhancing function of a Pd ectodomain.
Figure 7.
Pd exclusion is dominant over Pd localization. Subcellular localization is shown for chimeric fusions between AGP4 and PDCB1 in transient expression in N. benthamiana. A to C, Localization of SP-mCitrine fused to AGP4 in which the GPI signal was replaced with that of PDCB1 (mCit-AGΔGPI-GPICB). D to F, Localization of SP-mCitrine fused to AGP4 in which the GPI signal was replaced with the full-length PDCB1 (mCit-AGΔGPI-CB). G to I, Localization of SP-mCitrine fused to PDCB1 in which the GPI signal was replaced with the full-length AGP4 (mCit-CBΔGPI-AG). A, D, and G show localization with no treatment; B to B′′, E to E′′, and H to H′′ show localization in plasmolyzed cells stained with FM4-64. In plasmolyzed cells, the dashed white line marks the cell wall, asterisks indicate the apoplastic space formed by the receding protoplast, and triangles indicate the position of the PM. C, F, and I show localization in the presence of the GPI biosynthesis inhibitor mannosamine. Bars = 10 µm.
DISCUSSION
In this work, we have shown that GPI modification serves as a signal to mediate the targeting of proteins to Pd. A comparative analysis of two classes of GPI-APs, Pd localized and non-Pd localized, revealed that the GPI modification signal in both groups functions as a primary Pd-sorting signal. However, the ectodomain acts downstream and either reinforces Pd accumulation (Pd proteins) or mediates sorting out from Pd (non-Pd proteins). The Pd-associated GPI-APs characterized here, BG_pap and PDCB1, localize to Pd (Figs. 2 and 3) and regulate Pd permeability by changing callose levels at the cell wall domain of Pd (Levy et al., 2007; Simpson et al., 2009). BG_pap belongs to a large family of cell wall enzymes sharing a GH17 hydrolytic domain that was shown to specifically recognize callose (β-1,3-glucan polymer) and catalyze its endohydrolysis (Hrmova and Fincher, 2009). PDCB1 carries a CBM43 functional domain that is found in various cell wall proteins and was shown to specifically bind callose (Barral et al., 2005; Simpson et al., 2009). Inhibition of the GPI modification pathway by mannosamine blocked the Pd targeting of both proteins and led to intracellular accumulation in the ER-like and vesicle-like structures (Fig. 2), similar to the effect produced in mammalian and plant cells (Lisanti et al., 1991; Pan et al., 1992; Sevlever and Rosenberry, 1993; De Caroli et al., 2011). Importantly, the expression of BG_pap and PDCB1 without their C-terminal regions, which include the GPI-anchoring signal, blocked their Pd targeting and led to predominant ER localization (Fig. 2), indicating that the functional modules GH17BG and CBM43CB require membrane anchoring for Pd delivery. Recently, Grison et al. (2015) showed that mCherry-tagged PDCB1 without the GPI signal, but with its C-terminal unstructured region, had a soluble apoplastic localization pattern with no Pd enrichment, suggesting that the unstructured region in PDCB1 might have a role in enhancing the export of CBM43CB from the ER. Similar effects were shown with other plant GPI-APs, in which deletion of the GPI signal resulted either in a complete lack of protein export (Lee et al., 2009; Li et al., 2015; Deng et al., 2016) or soluble extracellular accumulation with no PM fraction (Sun et al., 2004; Li et al., 2010). Interestingly, all Pd-associated GH17 and CBM43 proteins reported to date are predicted GPI-APs (Levy et al., 2007; Simpson et al., 2009; Rinne et al., 2011; Benitez-Alfonso et al., 2013; Gaudioso-Pedraza and Benitez-Alfonso, 2014). These data, together with our findings, clearly show that GPI modification is a necessary posttranslational modification for delivering GH17 and CBM43 modules to Pd. However, not all predicted GPI-anchored GH17 and CBM43 proteins target to Pd. A recent bioinformatic analysis of putative GPI-anchored GH17 proteins from Arabidopsis revealed two groups of callose-modifying proteins: Pd localized and non-Pd localized (Gaudioso-Pedraza and Benitez-Alfonso, 2014). Members of the non-Pd group containing the GH17 and CBM43 modules showed soluble extracellular accumulation despite the presence of a GPI signal and affinity of their functional domains to callose substrate (Rinne et al., 2011; Gaudioso-Pedraza and Benitez-Alfonso, 2014). These observations, together with our analysis of the non-Pd GPI-APs, suggest that signals other than GPI modification and callose recognition determine the non-Pd localization in those proteins.
A significant observation made from this work is that the C-terminal GPI modification signal alone is able to target a reporter protein to Pd (Fig. 3). Moreover, the Pd-targeting function of the GPI signal was independent of its source, as signals from both Pd and non-Pd GPI-APs were able to mediate the Pd localization of mCitrine (Fig. 4; Supplemental Fig. S3). Previously published GPI-anchored fluorescent proteins in plant cells did not show a Pd-enriched accumulation but rather focused on the general PM localization of these reporters (Martinière et al., 2012; Baral et al., 2015). The GFP-GPIAGP4 fusion reported by Martinière et al. (2012) was created by fusing SP-GFP to Ser-107 of the AGP4 sequence, which is a lower score omega site compared with the higher score Ser-111 used for the SP-mCitrine fusion in this work (Fig. 1). Fusion of SP-mCitrine to Ser-107 indeed resulted in a reduced Pd accumulation and led to a stronger PM localization with the appearance of vesicle-like structures (Supplemental Fig. S5), suggesting that the residual four amino acids function as part of the ectodomain and have a Pd exclusion effect. The GFP-GPICOBRA and mCherry-GPICOBRA fusions reported by Baral et al. (2015) showed a strong PM localization with some intracellular vesicular structures in young root epidermal cells. Lack of Pd enrichment could be explained by high levels of fluorescent protein accumulation in the young root cells coupled with conditions of high exposure and gain, both of which would increase the background PM signal and make Pd enrichment impossible to distinguish. We observed enriched Pd localization for mCit-GPICOBRA in our expression system using leaf epidermal cells (Supplemental Fig. S6). We also tested the localization of SP-GFP-tagged GPI signal and observed a similar Pd enrichment (Supplemental Fig. S6), indicating that the effect is not limited to the mCitrine tag. Taken together, these data show that the different GPI signals characterized here function similarly in mediating Pd targeting and suggest that they allow attachment of the same GPI anchor to the proteins. However, we cannot exclude the possibility that some GPI signals may function differently in mediating the GPI modification and Pd targeting of a reporter protein. Analysis of GPI signals from additional groups of GPI-APs is required to determine their Pd-targeting ability and possibly to elucidate motif(s) within the GPI signal that may control its Pd-targeting function.
The Pd-enriched localization of GPI-anchored reporter protein likely represents its accumulation in the specialized regions of the PM, termed lipid rafts or PM microdomains. It was found recently that the region of the PM that lines the Pd channel is highly enriched in sphingolipids and sterols (Grison et al., 2015). Moreover, it was shown that Pd localization of GPI-APs, like PDCB1 and PDBG2, could be disrupted by inhibiting the sterol biosynthesis pathway (Grison et al., 2015). In addition, proteomic analysis of sterol-enriched detergent-resistant membranes in Arabidopsis revealed that GPI-APs are overrepresented in those membranes (Kierszniowska et al., 2009). Indeed, more detailed studies in mammalian cells show that an association with membrane microdomains is a general property of GPI-APs; however, the pattern of their lateral organization at the cell surface is determined by additional signals associated with protein ectodomain, cell type, and physiological state (Muñiz and Zurzolo, 2014; Paladino et al., 2015; Saha et al., 2016). Expression of BG_pap and PDCB1 with their ectodomains resulted in a higher accumulation at Pd, but not at the PM, compared with GPI-anchored mCitrine (Fig. 5). Similar levels of accumulation at the PM suggest that full-length proteins and GPI-anchored mCitrine have similar rates of secretion and targeting to the PM, including the PM domain of Pd. However, additional enrichment of the full-length proteins at Pd possibly results from interaction of the ectodomains with Pd-associated callose. Such interaction may occur through callose recognizing GH17BG and CBM43CB functional modules and may facilitate release of the proteins from the PM, enabling enriched accumulation in the cell wall domain of Pd. Supportive evidence for such a mode of accumulation was observed under stress conditions triggered by SA, where a complete release from the PM did not affect the Pd fraction of the full-length protein but eliminated that of the GPI-anchored mCitrine (Supplemental Fig. S4). We conclude that BG_pap and PDCB1 have two phases in their Pd localization: (1) GPI-mediated targeting to the PM domain of Pd and (2) ectodomain-mediated accumulation in the cell wall domain of Pd. We predict that, for other Pd-associated GPI-APs that function at the PM domain of Pd, like LYM2 (Faulkner et al., 2013), the Pd stabilization may occur through interaction of their ectodomain with a Pd-enriched transmembrane protein. In the case of LYM2, such a protein would be the FLAGELLIN SENSING2 transmembrane receptor that was shown to be enriched at Pd (Faulkner et al., 2013).
Unlike Pd-associated GPI-APs, the ectodomains in non-Pd GPI-APs abolished the Pd-targeting function of their own GPI signals as well as the GPI signal from a Pd protein (Figs. 6 and 7). Interestingly, despite significant PM fraction in both AGP4 and LTPG1, the proteins did not show Pd enrichment (Fig. 6). Previously reported non-Pd GPI-APs with exclusive PM localization also did not show Pd enrichment (Debono et al., 2009; Zhang et al., 2011a, 2011b; Li et al., 2013, 2015). Together, these observations suggest that Pd exclusion mediated by a non-Pd ectodomain likely occurs prior to arrival at the PM and Pd, possibly at the level of vesicle sorting in Golgi membranes. An alternative possibility would be that non-Pd GPI-APs are excluded from Pd at the PM, due to extensive cleavage of the GPI anchor. However, the latter possibility is unlikely, since a reciprocal fusion between Pd and non-Pd ectodomains resulted in an exclusive non-Pd localization (Fig. 7), which again indicates that Pd exclusion most likely occurs prior to PM and Pd localization. Studies in mammalian cells have shown that the sorting of different groups of GPI-APs occurs at the trans-Golgi network (TGN; Muñiz and Zurzolo, 2014). In polarized human and Madin-Darby canine kidney epithelial cells, the GPI-APs that target to the microdomain-rich apical PM segregate from the basolateral GPI-APs by forming large microdomain-rich clusters at the TGN (Muñiz and Zurzolo, 2014). It was shown that the apical GPI-APs indeed arrive at the PM as immobile clusters that form in the Golgi network (Hannan et al., 1993). A similar clustering mechanism taking place in the Golgi membranes may function in determining the Pd targeting of GPI-APs in plants. Our data suggest that accumulation in the Pd-destined membrane microdomains is a primary pathway for a GPI-AP and that the non-Pd GPI-APs are sorted out from this pathway by signals associated with their ectodomains. Such signals could be glycosylations at specific residues and/or interactions of the ectodomains with specific vesicle-sorting determinants at the TGN. However, as with mammalian cells (Paladino et al., 2008), it is possible that, for some plant GPI-APs, the GPI modification and remodeling steps also may contribute to correct sorting in the secretory pathway.
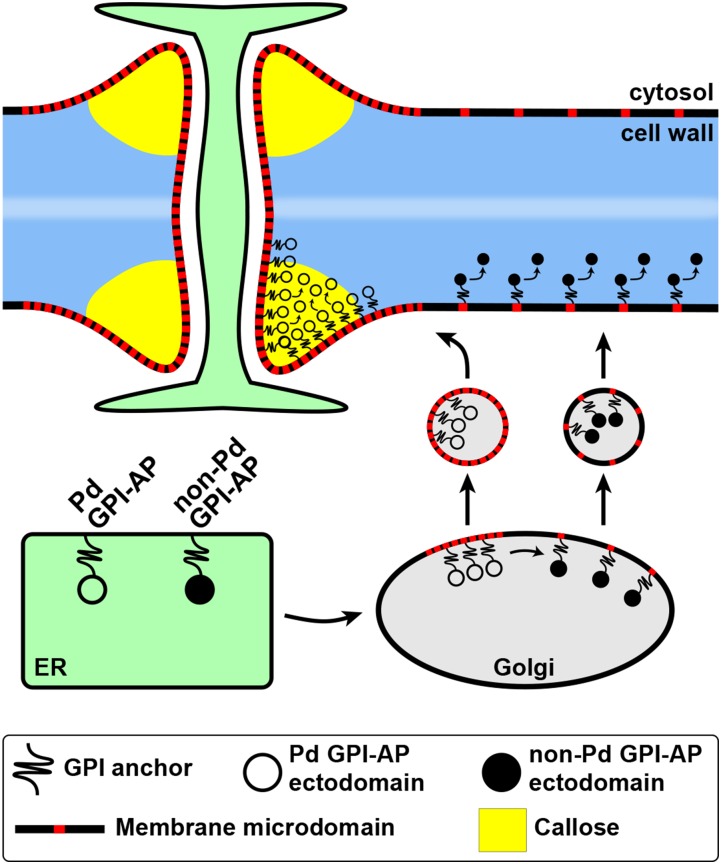
Based on our findings, we propose a model that describes the targeting of Pd-associated callose-modifying and non-Pd GPI-anchored proteins in plant cells (Fig. 8). Future studies will focus on determining signals that control the segregation and sorting of different GPI-APs during their targeting to specific regions of the PM and the cell wall. The distinct Pd-enriched localization of GPI-anchored reporters can serve as a sensitive molecular tool in studying the composition and functioning of the Pd-associated membrane microdomains as well as uncovering the complex mechanism of GPI-AP sorting in plant cells.
Figure 8.
Hypothetical model describing the targeting of callose-modifying Pd-associated GPI-anchored proteins (Pd GPI-APs) and non-Pd GPI-APs. After attachment of the GPI anchor (black string) in the ER and its subsequent remodeling (not shown), both groups of GPI-APs enter the secretory pathway. The remodeled GPI-APs partition into specialized membrane microdomains (red) in the Golgi membranes. The Pd-associated GPI-APs are sorted into microdomain-rich clusters, whereas the non-Pd GPI-APs are sorted out from such clusters by means of signals associated with their ectodomains. Vesicles formed from respective regions of the Golgi membranes deliver the GPI-APs to the PM, either to microdomain-rich regions like Pd (Pd GPI-APs) or other regions (non-Pd GPI-APs). The Pd-associated callose-modifying GPI-APs accumulate in the cell wall domain of Pd through interaction of their ectodomains with a Pd component (e.g. callose). The non-Pd GPI-APs are distributed uniformly at the cell surface both as membrane-anchored and as released cell wall proteins.
MATERIALS AND METHODS
Plant Growth Conditions and Transformation
Arabidopsis (Arabidopsis thaliana Columbia-0) and Nicotiana benthamiana plants were grown under long-day conditions (16-h-light/8-h-dark cycle) at 22°C and 25°C, respectively. For transient expression, mature leaves of 4-week-old N. benthamiana plants were infiltrated with Agrobacterium tumefacienes strain GV3101 (optical density at 600 nm = 0.2) harboring appropriate plasmids, and gene expression was assessed at 48 h post infiltration. For stable expression in Arabidopsis, a floral dipping method (Clough and Bent, 1998) was used.
Construction of Plant Expression Plasmids
Full-length genes or gene segments without the native N-terminal SP were amplified from Arabidopsis cDNA using gene-specific primers. Fusions with mCitrine (Zacharias et al., 2002) were constructed using SP taken from the Arabidopsis chitinase gene (At3g12500) and expressed under the control of the 35S promoter. First, pTZ57-SP-mCitrine plasmid was created by inserting the 30-amino acid SP sequence upstream of and in frame with the mCitrine coding region, between XbaI and EcoRI sites. The BG_pap and AGP4-related SP-mCitrine fusions were created by traditional cloning, as follows. The appropriate gene segment with the stop codon was amplified from cDNA and inserted into pTZ57-SP-mCitrine, downstream of and in frame with SP-mCitrine between KpnI and NotI sites. Then, the expression cassette SP-mCitrine-gene of interest was confirmed by sequencing and transferred into the binary plasmid pBIN-GFP (Sagi et al., 2005) between XbaI and NotI sites by replacing the GFP gene. The PDCB1- and LTPG1-related SP-mCitrine fusions, as well as the chimeric fusions between PDCB1 and AGP4, were created similarly, except using overlap PCR and subsequent Gateway technology (Invitrogen). First, the overlap PCR product SP-mCitrine-gene of interest was recombined into pDONR207 (Invitrogen) using AttB sites to create entry vectors. After sequencing, the entry vectors were recombined into pLN462 binary plasmid (Jamir et al., 2004). The SP-mCitrine fusions were created with the following protein segments of a corresponding GPI-AP (for protein diagrams, see Fig. 1). For BG_pap-related fusions, mCit-BG, segment 27 to 425; mCit-BGΔC, segment 27 to 348; mCit-GPIBG, segment 401 to 425; mCit-TMBG, segment 408 to 425. For PDCB1-related fusions, mCit-CB, segment 20 to 201; mCit-CBΔC, segment 20 to 104; mCit-GPICB, segment 172 to 201. For AGP4-related fusions, mCit-AG, segment 22 to 135; mCit-GPIAG, segment 111 to 135; mCit-TMAG, segment 117 to 135. For LTPG1-related fusions, mCit-LT, segment 33 to 193; mCit-GPILT, segment 160 to 193. For chimeric fusions between PDCB1 and AGP4, mCit-AGΔGPI-GPICB, segment 22 to 110 of AGP4 and segment 172 to 201 of PDCB1; mCit-AGΔGPI-CB, segment 22 to 110 of AGP4 and segment 20 to 201 of PDCB1; mCit-CBΔGPI-AG, segment 20 to 171 of PDCB1 and segment 22 to 135 of AGP4. sec-mCitrine was created by amplifying the SP-mCitrine sequence with the stop codon from pTZ57-SP-mCitrine. The PM marker mCit-AGG2 was created by fusing mCitrine to the N terminus of AGG2 using overlap PCR. Primers used for the construction of each fusion are listed in Supplemental Table S1. MP-RFP (Boutant et al., 2010) and mCherry-ER (Nelson et al., 2007) were expressed from previously published constructs.
The protein domains were predicted with SMART (Schultz et al., 1998). The C-terminal GPI modification signal was predicted with the big-PI Plant Predictor tool (Eisenhaber et al., 2003). The transmembrane segment of the GPI signal was predicted with ΔG Prediction Server version 1.0 (Hessa et al., 2007).
Chemical Treatments, Plasmolysis, and Histochemical Staining
Mannosamine (d-mannosamine hydrochloride; Sigma) was applied by infiltration of 10 mm water solution 24 h after agroinfiltration. After another 24 h, the infiltrated area was sampled for microscopy. Treatment with SA was performed as described previously (Zavaliev et al., 2013): leaves were sprayed with 5 mm water solution of SA (Sigma) 24 h after agroinfiltration. Leaf tissue was sampled for microscopy 24 h after SA spray. Plasmolysis was performed by incubating leaf samples in 0.75 m mannitol solution. The PM was stained with a 1:2,000 dilution of 10 μg μL−1 stock solution of FM4-64 (Molecular Probes) prior to plasmolysis.
Protein Extraction and Immunoblotting
Total proteins were extracted from 100 mg of leaf tissue of N. benthamiana 48 h after agroinfiltration. Samples were homogenized in 300 µL of ice-cold extraction buffer (50 mm phosphate buffer, pH 7.2, 150 mm NaCl, 2 mm MgCl2, 40 μm MG115, 1% Triton X-114, and a cocktail of protease inhibitors [−EDTA; Roche]) and centrifuged 15 min at 14,000 rpm at 4°C. The supernatant was used as the total protein fraction. Proteins were resolved with 4% to 12% Bis-Tris SDS-PAGE and probed with anti-GFP monoclonal antibody (Clontech).
Microscopy
Confocal fluorescence microscopy was performed with a Zeiss LSM 510 inverted confocal microscope using a 40×/1.3 oil-immersion objective. mCitrine was excited with an argon laser using a 514-nm beam splitter, and emission was detected with a 520- to 555-nm band pass filter. For colocalization with red fluorescence, mCitrine was excited using a 488-nm beam splitter, and emission was detected with a 505- to 550-nm band pass filter. RFP, mCherry, and FM4-64 were excited with a 561-nm diode laser, and emission was detected with a 575- to 615-nm band pass filter. The Pd- and PM-associated mCitrine fluorescence was quantified from at least 15 randomly sampled nonsaturated confocal images (512 × 512 pixels, 225 × 225 μm) using ImageJ. In each image, intensities from 15 to 20 Pd foci and adjacent PM regions were quantified using the same size area.
Accession Numbers
Accession numbers for the genes characterized in this work are as follows: BG_pap (At5g42100), PDCB1 (At5g61130), AGP4 (At5g10430), LTPG1 (At1g27950), COBRA (At5g60920), AGG2 (At3g22942), and BASIC CHITINASE (At3g12500).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Western-blot analysis of the fusion proteins characterized in this study.
Supplemental Figure S2. Localization of SP-mCitrine fused to the transmembrane segment of the GPI signal.
Supplemental Figure S3. Localization of Pd- and non-Pd GPI-APs in transgenic Arabidopsis.
Supplemental Figure S4. Effects of SA-induced stress on the localization of mCit-BG and mCit-GPIBG.
Supplemental Figure S5. Subcellular localization of SP-mCitrine fused to full-length and truncated AGP4.
Supplemental Figure S6. Subcellular localization of SP-mCitrine fused to GPI signal of Arabidopsis COBRA protein and SP-GFP fused to GPI signal of BG_pap.
Supplemental Table S1. List of primers used for cloning.
Supplementary Material
Acknowledgments
We thank Manfred Heinlein for providing us with MP-RFP clone, Yangnan Gu for mCherry-ER clone, and John Withers for critical reading of the article.
Glossary
- Pd
plasmodesmata
- PM
plasma membrane
- GPI
glycosylphosphatidylinositol
- GPI-AP
glycosylphosphatidylinositol-anchored protein
- SP
signal peptide
- ER
endoplasmic reticulum
- SA
salicylic acid
- TGN
trans-Golgi network
Footnotes
This work was supported by the Israel Science Foundation (grant no. 723/00–17.1 to B.L.E.), the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (grant no. GBMF3032 to X.D.), and a Vaadia-BARD Postdoctoral Fellowship (award no. FI–477–2013 to R.Z.) from BARD, The United States-Israel Binational Agricultural Research and Development Fund.
Articles can be viewed without a subscription.
References
- Ambrose C, DeBono A, Wasteneys G (2013) Cell geometry guides the dynamic targeting of apoplastic GPI-linked lipid transfer protein to cell wall elements and cell borders in Arabidopsis thaliana. PLoS ONE 8: e81215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral A, Irani NG, Fujimoto M, Nakano A, Mayor S, Mathew MK (2015) Salt-induced remodeling of spatially restricted clathrin-independent endocytic pathways in Arabidopsis root. Plant Cell 27: 1297–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral P, Suárez C, Batanero E, Alfonso C, de Dios Alché J, Rodríguez-García MI, Villalba M, Rivas G, Rodríguez R (2005) An olive pollen protein with allergenic activity, Ole e 10, defines a novel family of carbohydrate-binding modules and is potentially implicated in pollen germination. Biochem J 390: 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EM, Mongrand S, Tilsner J (2014) Specialized membrane domains of plasmodesmata, plant intercellular nanopores. Front Plant Sci 5: 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Faulkner C, Pendle A, Miyashima S, Helariutta Y, Maule A (2013) Symplastic intercellular connectivity regulates lateral root patterning. Dev Cell 26: 136–147 [DOI] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P (2002) Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a genomic analysis. Plant Physiol 129: 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutant E, Didier P, Niehl A, Mély Y, Ritzenthaler C, Heinlein M (2010) Fluorescent protein recruitment assay for demonstration and analysis of in vivo protein interactions in plant cells and its application to Tobacco mosaic virus movement protein. Plant J 62: 171–177 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Debono A, Yeats TH, Rose JK, Bird D, Jetter R, Kunst L, Samuels L (2009) Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21: 1230–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Caroli M, Lenucci MS, Di Sansebastiano GP, Dalessandro G, De Lorenzo G, Piro G (2011) Protein trafficking to the cell wall occurs through mechanisms distinguishable from default sorting in tobacco. Plant J 65: 295–308 [DOI] [PubMed] [Google Scholar]
- Deng T, Yao H, Wang J, Wang J, Xue H, Zuo K (2016) GhLTPG1, a cotton GPI-anchored lipid transfer protein, regulates the transport of phosphatidylinositol monophosphates and cotton fiber elongation. Sci Rep 6: 26829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber B, Wildpaner M, Schultz CJ, Borner GHH, Dupree P, Eisenhaber F (2003) Glycosylphosphatidylinositol lipid anchoring of plant proteins: sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol 133: 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M, Egelund J, Schultz CJ, Bacic A (2010) Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol 153: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ (2013) LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci USA 110: 9166–9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson MAJ, Kinoshita T, Hart GW (2009) Glycosylphosphatidylinositol anchors. In Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, eds, Essentials of Glycobiology, Ed 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Frigerio L, Jolliffe NA, Di Cola A, Felipe DH, Paris N, Neuhaus JM, Lord JM, Ceriotti A, Roberts LM (2001) The internal propeptide of the ricin precursor carries a sequence-specific determinant for vacuolar sorting. Plant Physiol 126: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudioso-Pedraza R, Benitez-Alfonso Y (2014) A phylogenetic approach to study the origin and evolution of plasmodesmata-localized glycosyl hydrolases family 17. Front Plant Sci 5: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda SK, Shockey JM, Rothstein SJ, Dyer JM, Mullen RT (2009) Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiol Biochem 47: 867–879 [DOI] [PubMed] [Google Scholar]
- González-Solís A, Cano-Ramírez DL, Morales-Cedillo F, Tapia de Aquino C, Gavilanes-Ruiz M (2014) Arabidopsis mutants in sphingolipid synthesis as tools to understand the structure and function of membrane microdomains in plasmodesmata. Front Plant Sci 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grison MS, Brocard L, Fouillen L, Nicolas W, Wewer V, Dörmann P, Nacir H, Benitez-Alfonso Y, Claverol S, Germain V, et al. (2015) Specific membrane lipid composition is important for plasmodesmata function in Arabidopsis. Plant Cell 27: 1228–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan LA, Lisanti MP, Rodriguez-Boulan E, Edidin M (1993) Correctly sorted molecules of a GPI-anchored protein are clustered and immobile when they arrive at the apical surface of MDCK cells. J Cell Biol 120: 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G (2007) Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hrmova M, Fincher GB (2009) Plant and microbial enzymes involved in the depolymerization of (1,3)-beta-D-glucans and related polysaccharides In Bacic A, Fincher GB, Stone BA, eds, Chemistry, Biochemistry and Biology of (1-3)-beta-Glucans and Related Polysaccharides. Academic Press, New York, pp 119–170 [Google Scholar]
- Jamir Y, Guo M, Oh HS, Petnicki-Ocwieja T, Chen S, Tang X, Dickman MB, Collmer A, Alfano JR (2004) Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J 37: 554–565 [DOI] [PubMed] [Google Scholar]
- Kierszniowska S, Seiwert B, Schulze WX (2009) Definition of Arabidopsis sterol-rich membrane microdomains by differential treatment with methyl-beta-cyclodextrin and quantitative proteomics. Mol Cell Proteomics 8: 612–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T. (2015) Structural changes of GPI anchor after its attachment to proteins: functional significance. In Chakrabarti A, Surolia A, eds, Biochemical Roles of Eukaryotic Cell Surface Macromolecules. Springer-Verlag, New York, pp 17–25 [DOI] [PubMed] [Google Scholar]
- Knox JP, Benitez-Alfonso Y (2014) Roles and regulation of plant cell walls surrounding plasmodesmata. Curr Opin Plant Biol 22: 93–100 [DOI] [PubMed] [Google Scholar]
- Lee SB, Go YS, Bae HJ, Park JH, Cho SH, Cho HJ, Lee DS, Park OK, Hwang I, Suh MC (2009) Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola. Plant Physiol 150: 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Erlanger M, Rosenthal M, Epel BL (2007) A plasmodesmata-associated β-1,3-glucanase in Arabidopsis. Plant J 49: 669–682 [DOI] [PubMed] [Google Scholar]
- Li C, Yeh FL, Cheung AY, Duan Q, Kita D, Liu MC, Maman J, Luu EJ, Wu BW, Gates L, et al. (2015) Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yu M, Geng LL, Zhao J (2010) The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J 64: 482–497 [DOI] [PubMed] [Google Scholar]
- Li S, Ge FR, Xu M, Zhao XY, Huang GQ, Zhou LZ, Wang JG, Kombrink A, McCormick S, Zhang XS, et al. (2013) Arabidopsis COBRA-LIKE 10, a GPI-anchored protein, mediates directional growth of pollen tubes. Plant J 74: 486–497 [DOI] [PubMed] [Google Scholar]
- Lisanti MP, Field MC, Caras IW, Menon AK, Rodriguez-Boulan E (1991) Mannosamine, a novel inhibitor of glycosylphosphatidylinositol incorporation into proteins. EMBO J 10: 1969–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinière A, Lavagi I, Nageswaran G, Rolfe DJ, Maneta-Peyret L, Luu DT, Botchway SW, Webb SE, Mongrand S, Maurel C, et al. (2012) Cell wall constrains lateral diffusion of plant plasma-membrane proteins. Proc Natl Acad Sci USA 109: 12805–12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñiz M, Zurzolo C (2014) Sorting of GPI-anchored proteins from yeast to mammals: common pathways at different sites? J Cell Sci 127: 2793–2801 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Paladino S, Lebreton S, Tivodar S, Campana V, Tempre R, Zurzolo C (2008) Different GPI-attachment signals affect the oligomerisation of GPI-anchored proteins and their apical sorting. J Cell Sci 121: 4001–4007 [DOI] [PubMed] [Google Scholar]
- Paladino S, Lebreton S, Zurzolo C (2015) Trafficking and membrane organization of GPI-anchored proteins in health and diseases. Curr Top Membr 75: 269–303 [DOI] [PubMed] [Google Scholar]
- Pan YT, Kamitani T, Bhuvaneswaran C, Hallaq Y, Warren CD, Yeh ET, Elbein AD (1992) Inhibition of glycosylphosphatidylinositol anchor formation by mannosamine. J Biol Chem 267: 21250–21255 [PubMed] [Google Scholar]
- Raffaele S, Bayer E, Lafarge D, Cluzet S, German Retana S, Boubekeur T, Leborgne-Castel N, Carde JP, Lherminier J, Noirot E, et al. (2009) Remorin, a Solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne PL, Welling A, Vahala J, Ripel L, Ruonala R, Kangasjärvi J, van der Schoot C (2011) Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-β-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 23: 130–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi G, Katz A, Guenoune-Gelbart D, Epel BL (2005) Class 1 reversibly glycosylated polypeptides are plasmodesmal-associated proteins delivered to plasmodesmata via the Golgi apparatus. Plant Cell 17: 1788–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Anilkumar AA, Mayor S (2016) GPI-anchored protein organization and dynamics at the cell surface. J Lipid Res 57: 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA 95: 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevlever D, Rosenberry TL (1993) Mannosamine inhibits the synthesis of putative glycoinositol phospholipid anchor precursors in mammalian cells without incorporating into an accumulated intermediate. J Biol Chem 268: 10938–10945 [PubMed] [Google Scholar]
- Sherrier DJ, Prime TA, Dupree P (1999) Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis 20: 2027–2035 [DOI] [PubMed] [Google Scholar]
- Simpson C, Thomas C, Findlay K, Bayer E, Maule AJ (2009) An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TK. (2009) Inhibitors of GPI biosynthesis. Enzymes 26: 247–267 [Google Scholar]
- Stahl Y, Grabowski S, Bleckmann A, Kühnemuth R, Weidtkamp-Peters S, Pinto KG, Kirschner GK, Schmid JB, Wink RH, Hülsewede A, et al. (2013) Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol 23: 362–371 [DOI] [PubMed] [Google Scholar]
- Sun W, Zhao ZD, Hare MC, Kieliszewski MJ, Showalter AM (2004) Tomato LeAGP-1 is a plasma membrane-bound, glycosylphosphatidylinositol-anchored arabinogalactan-protein. Physiol Plant 120: 319–327 [DOI] [PubMed] [Google Scholar]
- Thomas CL, Bayer EM, Ritzenthaler C, Fernandez-Calvino L, Maule AJ (2008) Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol 6: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J, Amari K, Torrance L (2010) Plasmodesmata viewed as specialised membrane adhesion sites. Protoplasma 248: 39–60 [DOI] [PubMed] [Google Scholar]
- Tilsner J, Nicolas W, Rosado A, Bayer EM (2016) Staying tight: plasmodesmal membrane contact sites and the control of cell-to-cell connectivity in plants. Annu Rev Plant Biol 67: 337–364 [DOI] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al. (2011) Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 21: 1144–1155 [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296: 913–916 [DOI] [PubMed] [Google Scholar]
- Zavaliev R, Levy A, Gera A, Epel BL (2013) Subcellular dynamics and role of Arabidopsis β-1,3-glucanases in cell-to-cell movement of tobamoviruses. Mol Plant Microbe Interact 26: 1016–1030 [DOI] [PubMed] [Google Scholar]
- Zavaliev R, Ueki S, Epel BL, Citovsky V (2011) Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma 248: 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Wang X, Running MP (2007) Dual lipid modification of Arabidopsis Gγ-subunits is required for efficient plasma membrane targeting. Plant Physiol 143: 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang J, Showalter AM (2011a) AtAGP18 is localized at the plasma membrane and functions in plant growth and development. Planta 233: 675–683 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang J, Showalter AM (2011b) AtAGP18, a lysine-rich arabinogalactan protein in Arabidopsis thaliana, functions in plant growth and development as a putative co-receptor for signal transduction. Plant Signal Behav 6: 855–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.