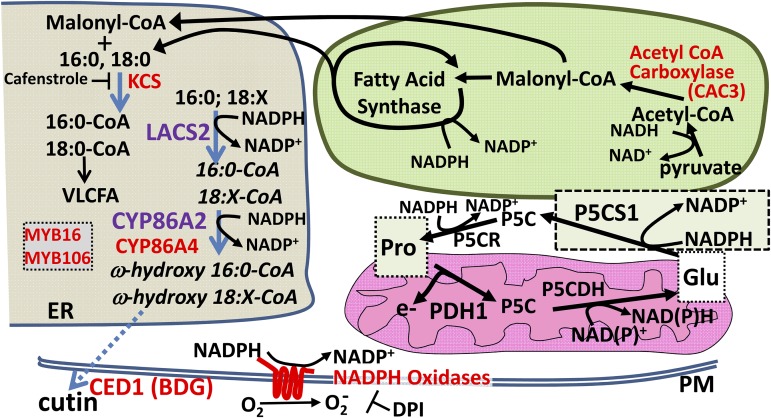
Figure 8.
Summary diagram showing the core pathway of Pro metabolism in relation to lipid, cuticle, and redox-related genes found to affect Pro accumulation. Multiple genes and pharmacological treatments disrupting lipid metabolism or NADPH oxidase activity were found to affect Pro accumulation and PDH1pro:LUC2 expression. This included CYP86A2 and LACS2 identified in the PDH1pro:LUC2 forward genetic screen; CYP86A4, CED1, MYB16, and MYB106 identified by reverse genetics; CAC3 and KCS genes identified by p5cs1-4 RNA sequencing; as well as strong effects of the NADPH oxidase inhibitor DPI. The reactions catalyzed by these proteins are spread across several compartments, including endoplasmic reticulum (ER), chloroplast, and plasma membrane (PM). The commonality among these pathways and Pro metabolism itself is an effect on redox status by the consumption of NADPH and the regeneration of NADP+, either directly, as for P5CS1, P5CR, LACS2, CYP86A2, and NADPH oxidases, or indirectly by modifying the flux through lipid synthesis, as for CAC3, KCS, CED1, MYB16, and MYB106. This commonality, along with data obtained with ROS scavengers and DTT treatment, indicates that redox status is a key factor linking these pathways together. Consistent with this, RNA sequencing data showed that p5cs1-4 had substantial alterations in gene expression-related chloroplast and mitochondrial redox metabolism, further indicating the relationship of Pro to redox status. The similar effects of p5cs1-4 on gene expression at both high and low ψw indicated the importance of Pro metabolism, and likely a high rate of flux through the Pro cycle of synthesis and catabolism, even in unstressed plants where Pro level is low. P5CS1 is drawn in its own box to indicate that its subcellular localization is unclear. How changes in redox state are communicated between these subcellular compartments is unknown.