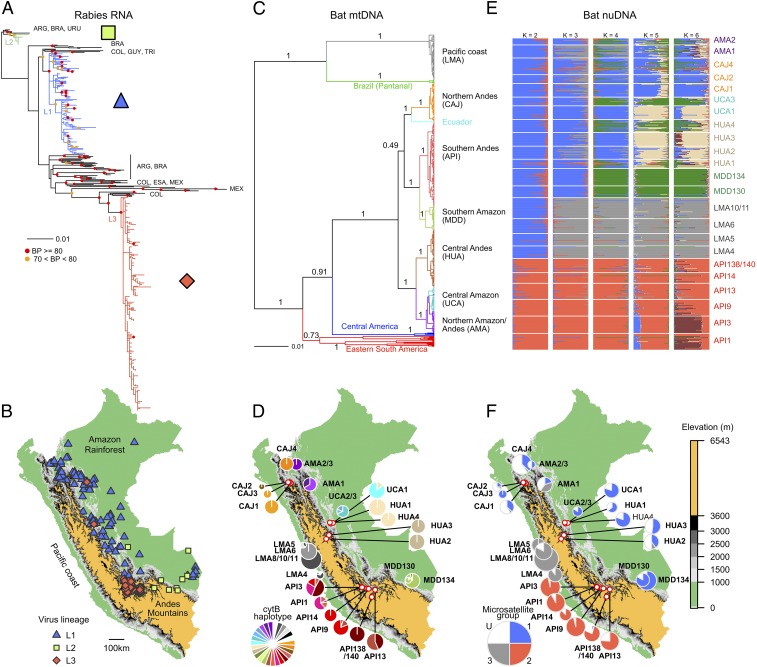
Fig. 1.
Genetic and geographic structure of host and viral markers with distinct inheritance mechanisms. (A) The ML tree of VBRV, using 434 complete N sequences from Peru (colored branches) and other representative countries in the Americas (black branches; ARG, Argentina; BRA, Brazil; COL, Colombia; ESA, El Salvador; GUY, French Guiana; MEX, Mexico; TRI, Trinidad; URU, Uruguay). Colored symbols show bootstrap support from 1,000 replicate ML searches. Two outgroup sequences from a rabies variant circulating in Peruvian dogs were excluded for visualization. (B) Geographic distributions of viral lineages in Peru. Areas above 3,600 m (the upper limit to vampire bats in Peru) are colored gold. (C) Bayesian phylogenetic tree of cytB sequences from vampire bats from Peru (n = 442) and other countries in the Americas (n = 26). Branches are colored by geographic region. Node values are posterior probabilities. (D) Distribution of CytB haplotypes across vampire bat colonies in Peru. Sites with <8 sequenced individuals were grouped with other colonies surveyed within 10 km. Pie charts are proportionate to sample size (range = 8–30). (E) Estimates from STRUCTURE analyses assuming K = 2–6 populations using 9 microsatellites (n = 480 bats). Each bar represents the probability of membership assignment to each of K groups. (F) Pie charts show the distribution of microsatellite groups [K = 3, threshold probability for group membership = 0.85, unassigned individuals (U) in white].