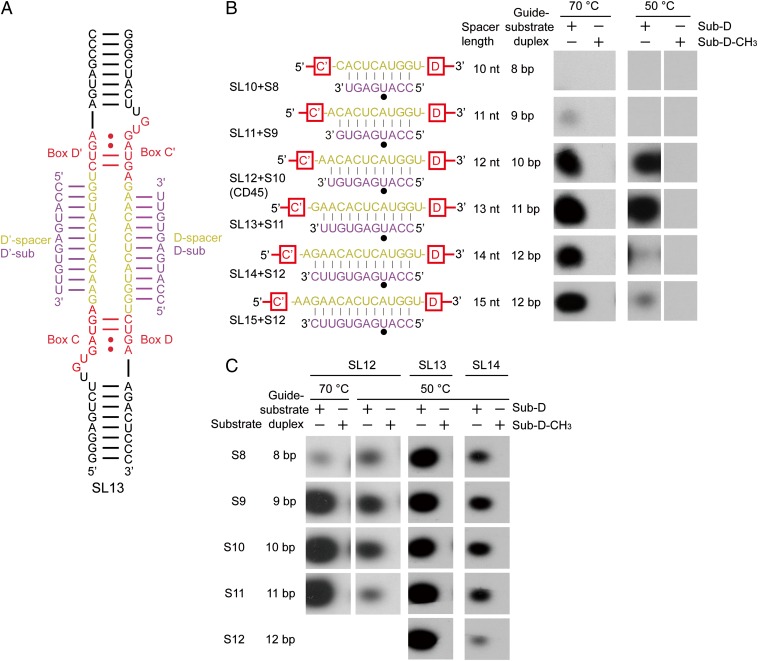
Fig. 1.
C/D RNP activities depend on the length of spacers and guide-substrate duplexes. (A) Secondary structure of the two-piece C/D RNA SL13. Other C/D RNAs differ only in the spacer regions. Boxes C, D, C′, and D′ are shown in red, the spacer regions are in yellow, and the bound substrates are in purple. (B and C) Activity of C/D RNPs. C/D RNPs were reconstituted with guide RNAs SL10–SL15 (1 μM) and incubated with substrates S8–S12 or their premethylated versions (30 μM), [methyl-3H] SAM, and cold SAM (30 μM) at 70 °C or 50 °C for 20 min. RNAs were separated by denaturing PAGE and visualized by 3H autoradiography. Base pairing interactions between the guide and the substrate are displayed. The target site is marked by a black circle.