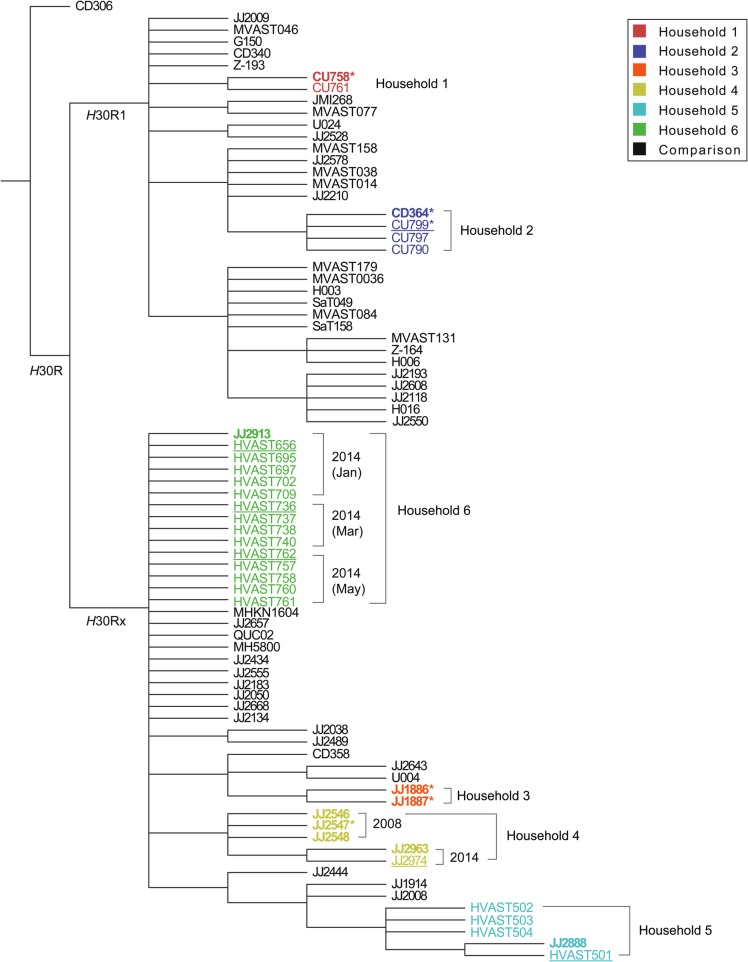
Figure 2.
Bootstrapped consensus core genome phylogeny for the 33 present household isolates and 47 comparison isolates of Escherichia coli sequence type 131 (ST131). The tree was based on 1000 bootstrapped maximum likelihood trees, retaining only those nodes that appeared in >70% of trees, and was rooted with strain CD306, which in the all-isolate phylogeny (data not shown) was basal. Clades H30R1 and H30Rx correspond, respectively, with clades C1 and C2 of Petty et al [8]. Branch lengths are meaningless. Household isolates are color-coded by household (1–6); comparison isolates, from Price et al [11], are shown in black. For the household isolates: boldface indicates clinical isolates; regular font indicates fecal isolates; underlining indicates fecal isolates from a clinical isolate's source host; and asterisks indicate the 6 household isolates that were included in Price et al [11], ie, JJ1886, JJ1887, JJ2547, CU758, CU799, and CD364 (which in Price et al was labeled as CD449). Dates are shown for the 2 households that underwent serial sampling.