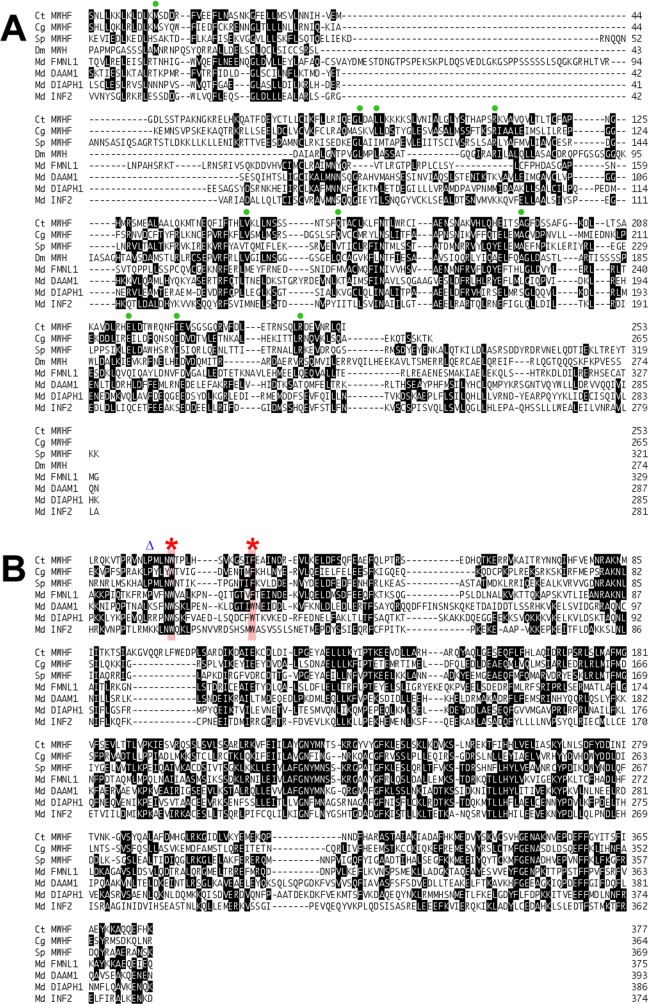
Fig 4. Sequence similarities between MWHF, MWH, and FMNL proteins.
(A) DID and DD sequences. Shown are a subset of amino acid sequences used to estimate the DID-DD phylogenetic tree of Fig 3B, including those from the MWHF proteins of the polychaete worm C. teleta (Ct), the Pacific oyster C. gigas (Cg), and the purple sea urchin S. purpuratus (Sp), MWH from D. melanogaster (Dm), and the formins FMNL1, DAAM1, DIAPH1, and INF2 from the opossum M. domestica (Md). Green circles indicate amino acid positions for which MWH and two or more MWHFs are identical but distinct from other formins. (B) FH2 domain sequences. Shown are sequences from the same subset of formins used to estimate the FH2 domain phylogenetic tree of Fig 1. Red asterisks indicate conserved aromatic residues of the FH2 domain lasso. The second aromatic residue is phenylalanine in FMNL and MWHF proteins, but tryptophan in all other formins. A blue triangle indicates a position uniquely occupied by proline in FMNL and MWHF proteins.