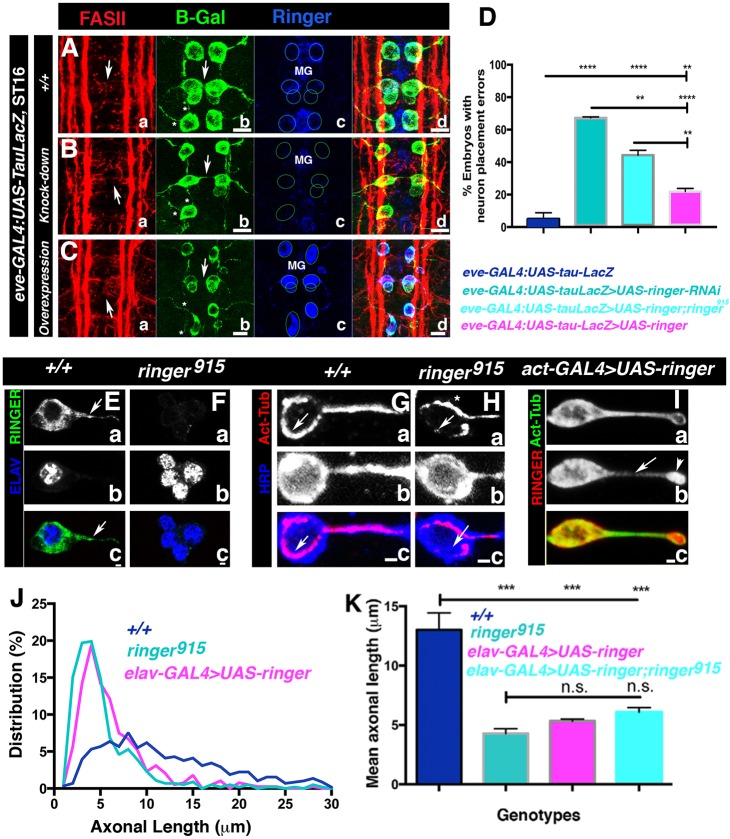
Fig. 4.
Changes in Ringer levels affect neuronal position and process extension. (A–C) FASII- (a), β-Galactosidase- (b, B-Gal) and Ringer-specific (c) immunostaining of eve-GAL4:UAS-tau-LacZ embryos shows arrangement of early stage-16 RP2, aCC and pCC neurons and their projections. (B) Ringer expression changes in Eve-positive neurons lead to axonal (A–C; arrows, disruption of cell placement; asterisks, axonal defects) and soma defects (A–C). For each genotype, neuron soma placement is shown by circles in Ac,Bc,Cc, and midline glia are MG. FASII-specific immunostaining shows axonal bundle disruption (Aa–Ca). (D) Quantification of neuronal misplacement (n=∼100). 67.25%±0.58 (mean±s.e.m.) of knockdown (P<0.0001) and 21.83%±1.94 of overexpressing (P=0.0082) embryos exhibited misplacement compared to 5%±3.8 in WT (ANOVA). Rescue experiments exhibited a modest reduction to 44.29%±2.98 compared to knockdown (P=0.0012) and WT (P<0.0001) (ANOVA). (E,F) Primary neuronal cultures from larval CNS show that Ringer was also expressed in axons (arrow) and absent in mutant cultures. (E–I) Neuron identity was determined by staining of ELAV (Db,Eb) or with an anti-HRP antibody (Fb,Gb). Scale bars: 1 μm. (G–I) Immunostaining for acetylated tubulin. In ringer915 (H) and overexpressing embryos (I) microtubules appear disorganized (arrowheads) compared to +/+ (G). Arrowhead in Ringer overexpression image (I) shows Ringer accumulation at the axon tip. Scale bar: 5 μm. (J) Percent frequency distribution of FASII-positive primary neurons up to 30 μm in length. Quantification included neurons with axons up to 100 μm in length that were positive for FASII, anti-HRP antibody binding and acetylated tubulin. (K) After 24 h, the mean axon length for +/+ embryos was 13 μm±1.4 compared to 4.3 μm±0.4 in mutants (P 0.0002). Overexpression using elav-GAL4>UAS-ringer in the WT or in the mutant background resulted in mean axonal lengths of 5.3 μm±2.7 (P=0.0005) and 6.1 μm±0.4 (P=0.0010), respectively (ANOVA). n.s., not significant; *P≤0.05, **P≤0.01. Error bars are s.e.m.