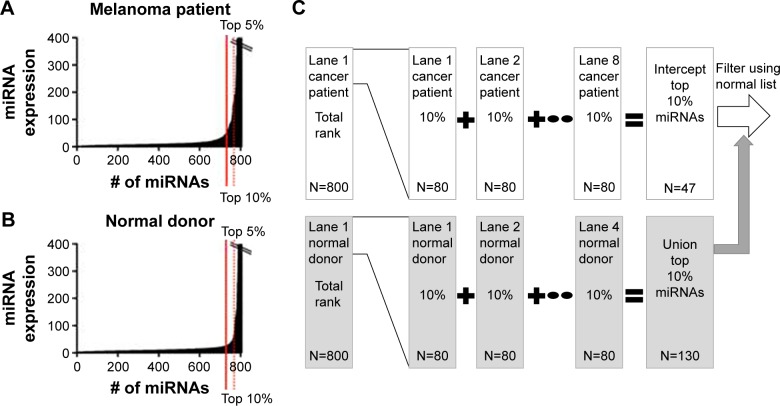
Figure 2.
Bioinformatics method to select upregulated microRNAs (miRNAs) for chip 1.
Notes: First, the 800 miRNAs identified by NanoString® in each lane (one lane per patient) were ordered from the highest to lowest expression (ranked). (A) A list of the top 5% (45) of the miRNAs present in the plasma of a cancer patient in chip 1 was generated. (B) A list of the top 5% (45) of the miRNAs that were present in the plasma of the first normal control donor was generated. (C) The process was repeated for each of the eight patient samples and four normal donors, and thus 12 such lists were created. The miRNAs that were present in any of the top 5% of the normal donor control lists (four lists, N=56; union of the control list) were removed from the miRNAs that were present in the top 5% of highly expressing miRNAs in all the patient lists (eight lists, N=25; intercept of the patient lists) to generate a list consisting of 22 upregulated miRNAs in patient samples. The selection of upregulated miRNAs begins in a convenient sample size of the top 5% of the miRNA list and repeats the process of finding upregulated miRNAs by expanding the feature list size (by 5%) through multiple iterations until the most number of upregulated miRNAs are selected that have a nonrandom false positive rate (estimated sensitivity and specificity >99.99%). For the first chip, the cutoff of selecting upregulated miRNAs is 10%. The upregulated miRNA final list (32 miRNAs) in this case is a combination of the miRNAs found to be upregulated in the top 5% or 10% lists (false positive rate <0.01).