(
A) To directly compare the nuclear level of Msn2-RFP relative to that of Msn4-YFP in the same single cells (
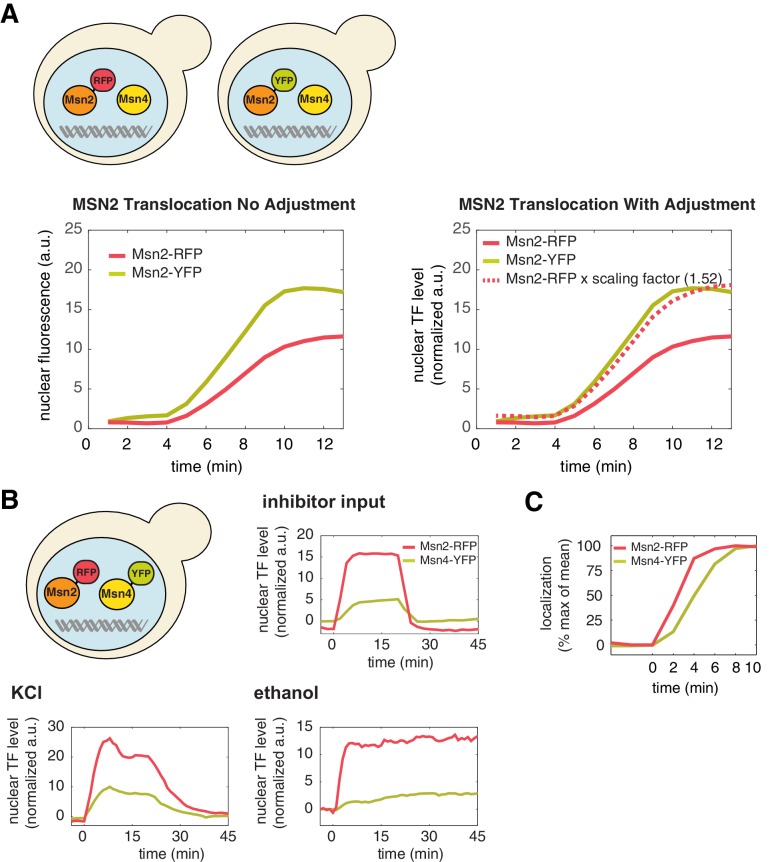
Figure 1A), a scaling factor between RFP and YFP is needed to account for unique microscope settings used in each channel as well as inherent emission differences between each fluorophore. This was determined by creating two yeast strains in which Msn2 was C-terminally tagged with either florescent protein RFP or YFP, respectively (illustrated in the top panel). Left: Sustained nuclear translocation of Msn2 was induced in both stains with an identical stimulus and the averaged single-cell time traces of Msn2 translocation were generated for both strains (n: ~100 cells per strain). Right: The scaling factor (1.52) was determined by taking the ratio between the maximal fluorescence intensity of each averaged trace. The time trace of Msn2-RFP, when times the scaling factor (dashed curve), overlaps with the time trace of Msn2-YFP. Therefore, this factor normalizes the fluorescence arbitrary unit of RFP with the fluorescence arbitrary unit of YFP and enables the direct comparison of the nuclear level of Msn2-RFP with that of Msn4-YFP in the same single cells (in the unit of 'normalized a.u.'). (
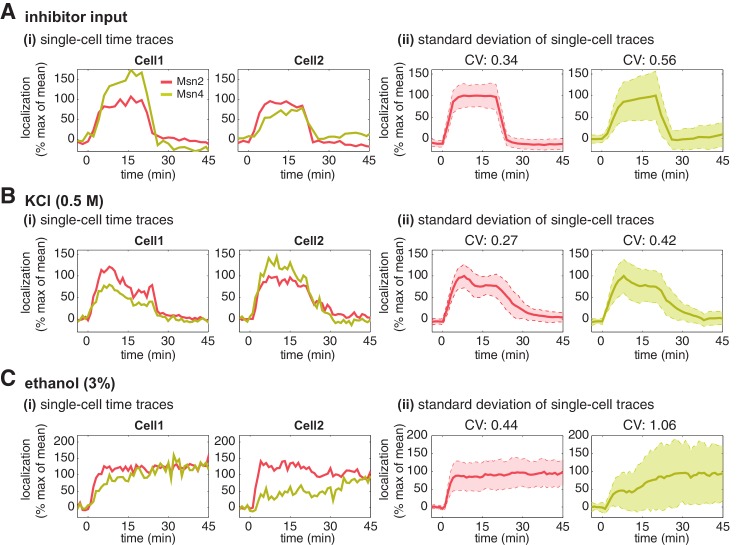
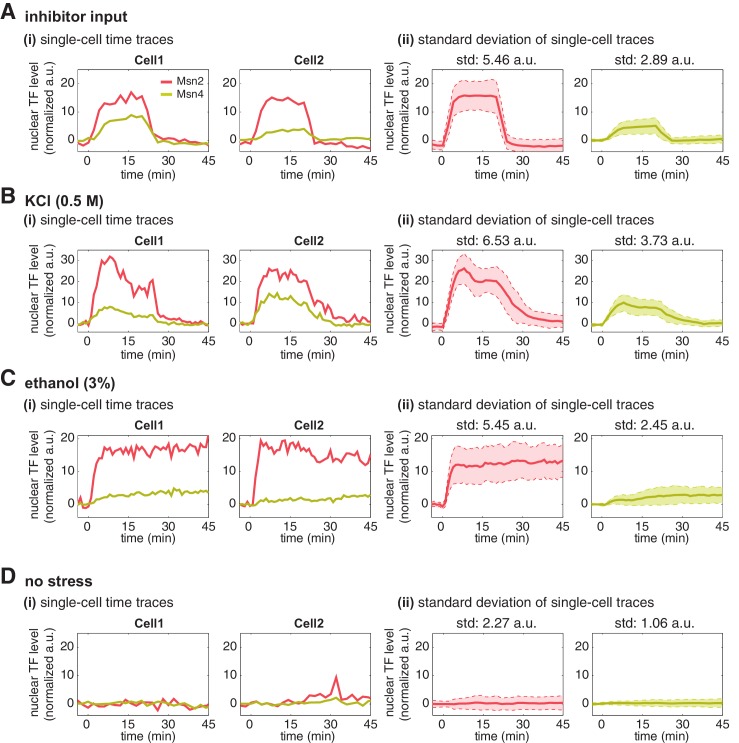
B) Averaged time traces of Msn2 and Msn4 nuclear translocation in the same single cells in response to 20-min 1 μM inhibitor pulse, 0.5M KCl, or 3% ethanol, as indicated. The top left panel illustrates that Msn2-YFP and Msn4-RFP are expressed in the same strain. The averaged traces were normalized by the scaling factor to allow a direct comparison of Msn2 and Msn4 in the same cells. (
C) Averaged time traces of Msn2 and Msn4 nuclear translocation from the same cells were normalized as% of max. The traces were plotted together and zoomed in to the early time period of the response to demonstrate the small time delay of Msn4 translocation.