Abstract
Background:
Inappropriate prescribing increases patient illness and death owing to adverse drug events. The inclusion of genetic information into primary care medication practices is one solution. Our aim was to assess the ability to obtain and genotype saliva samples and to determine the levels of use of a decision support tool that creates medication options adjusted for patient characteristics, drug-drug interactions and pharmacogenetics.
Methods:
We conducted a cohort study in 6 primary care settings (5 family practices and 1 pharmacy), enrolling 191 adults with at least 1 of 10 common diseases. Saliva samples were obtained in the physician's office or pharmacy and sent to our laboratory, where DNA was extracted and genotyped and reports were generated. The reports were sent directly to the family physician/pharmacist and linked to an evidence-based prescribing decision support system. The primary outcome was ability to obtain and genotype samples. The secondary outcomes were yield and purity of DNA samples, ability to link results to decision support software and use of the decision support software.
Results:
Genotyping resulted in linking of 189 patients (99%) with pharmacogenetic reports to the decision support program. A total of 96.8% of samples had at least 1 actionable genotype for medications included in the decision support system. The medication support system was used by the physicians and pharmacists 236 times over 3 months.
Interpretation:
Physicians and pharmacists can collect saliva samples of sufficient quantity and quality for DNA extraction, purification and genotyping. A clinical decision support system with integrated data from pharmacogenetic tests may enable personalized prescribing within primary care. Trial registration: ClinicalTrials.gov, NCT02383290.
Drug-related adverse events are an important and common cause of morbidity,1,2 with incidence rates as high as 25%.3 There is high-level evidence from systematic reviews and guidelines that pharmacogenetic testing is an effective method of reducing adverse drug events.4-6 Results from a US cohort of 1143 patients who experienced adverse events indicated that a clinically significant proportion of adverse effects, 33%, were due to drug-gene interactions or drug-drug-gene interactions.7 The Clinical Pharmacogenetics Implementation Consortium provides guidelines that enable the translation of genetic laboratory test results into actionable prescribing decisions for specific drugs.8 Of the approximately 4 billion prescriptions filled in the United States in 2013, 18% had actionable pharmacogenetics.9
Many of the drugs studied in pharmacogenetic trials are part of the primary care drug formulary and are used for common conditions. Pharmacogenetic panels are now available at an affordable price, and patients are requesting the tests and asking physicians to use these results in their care. Before a primary care pharmacogenetic panel is implemented, it is necessary to consider the ability of health care providers to incorporate this information into current medication selection processes.
Despite the increased use of electronic medical records, the prevalence of inappropriate (35%) and high-risk (14%) prescriptions remains high10-12 Alert systems have proven ineffective at changing prescribing decisions,13 whereas information given within a physician's workflow has been found to be effective in reducing inappropriate orders for imaging.14
A medication decision support system (MDSS) is a health information technology system that is designed to provide health care professionals with clinical decision support with medication decision-making tasks. These systems assess whether a drug is safe and effective for the patient, taking into account other medications, diseases and the patient's physical state.15 We have developed a patient-centred MDSS that assesses the potential drug-drug, drug-condition, drug-gene and drug-drug-gene interactions and produces a list of drug options least likely to cause harm and most likely to be effective.
We conducted a study to assess the DNA collection processes, investigate a panel of pharmacogenetic tests relevant to primary care patients and assess the use of an MDSS. The value of the MDSS will be assessed once feasibility of all processes has been shown.
Methods
Over an 18-month period before the start of the trial, we developed a pharmacogenetic panel, a pharmacogenetic report and an MDSS for use in primary care. This required custom software for pharmacogenetic report linking and running algorithms within the MDSS.
We used a prospective cohort study design, with family physicians and pharmacists recruiting patients between May and December 2015. Saliva samples were sent to our laboratory, where DNA was extracted and analyzed and reports were generated. Reports were sent directly to the family physician/pharmacist and linked to the MDSS. The physicians, pharmacists and support staff used the software to review the patient's therapy for 1 or more conditions and then provided feedback about the process.
Development of pharmacogenetic panel
We ranked single nucleotide polymorphisms and copy number variants according to clinical annotations primarily from PharmGKB,16 the Clinical Pharmacogenetic Implementation Consortium8 and the Royal Dutch Association for the Advancement of Pharmacy review.5 We selected a pharmacogenetic panel based on information from the PharmaADME Consortium (www.PharmaADME.org) as well as guidelines and drug labels (Appendix 1, available at www.cmajopen.ca/content/4/3/E528/suppl/DC1). Panel selection was by availability of validated assays from the manufacturer and consensus among authors (H.K.-E., M.A., S.A. and A.S.-G.) on the highest levels of evidence for clinical use in primary care. In addition, a custom assay for HLA-B*58:01 was developed and validated in house.
The selected panel included 33 of the top-ranking genetic variants in the following genes: CYP2C9, CYP2C19, CYP2D6, G6PD, HLA-B, SLCO1B1 and VKORC1. For clinical use, HLA-B tag/single nucleotide polymorphisms could not be used, and assay performance was low for 2 single nucleotide polymorphisms, resulting in a customized panel of 24 genetic variants for 20 drugs. We chose TaqMan allelic discrimination quantitative polymerase chain reaction assays for this study based on the manufacturer guarantee of working assays on the QuantStudio 12K Flex platform (Applied Biosystems).
We developed a quality-control and validation process to test the sensitivity and specificity of the assays. This included using Coriell Biorepository control samples, analyzing the results and comparing the experiment with known genotypes to determine feasibility and accuracy of the genetic test, and using sample replicates to assess concordance. In addition, we confirmed a subset of experimentally determined genotypes using Sanger sequencing. Once validated, we incorporated these assays onto our primary care pharmacogenetic panel.
Development of medication decision support system
We performed a literature review to find the most prevalent diseases in primary care populations, the diseases seen most frequently in family physicians' offices and the diseases treated by medications with an actionable pharmacogenetic test (Appendix 1). The 10 diseases that met the criteria and were selected were gout, chronic obstructive pulmonary disease, migraine, depression, osteoarthritis, hypertension, hyperlipidemia, atrial fibrillation, osteoporosis and epilepsy. These 10 conditions account for 15% of all primary care consultations, both medical and administrative.17 As 5 of the diseases have 2 distinct therapeutic approaches, the 10 diseases result in 15 "conditions." For example, the therapeutic options are very different for a patient with an acute flare-up of gout than for a patient with the chronic stages of gout.
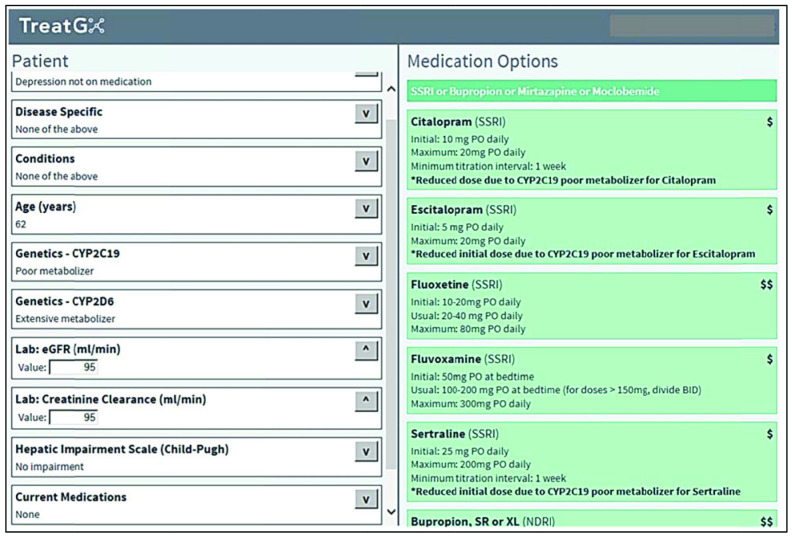
After identifying published epidemiological evidence for the management of a given condition, our team, consisting of pharmacists, physicians and experts in evidence-informed health care, used 1) the highest levels of evidence for treatment selection, 2) known drug-drug interactions from standard databases, published studies and product monographs and 3) pharmacogenetic information from PharmGKB (level 1A or 1B annotation in cases in which there were clear drug dosing guidelines) and other resources to form logic trees. All logic trees were tested extensively with the use of deidentified patient data obtained from 1 clinic's electronic health records (for which ethics approval was obtained) and were reviewed by our team of pharmacists and physicians. Logic trees were developed and tested by D.D, M.D., G.L., A.P. and P.S., alongwith 2 family physicians and a pharmacist. From the logic trees, we wrote the rules necessary for programming the MDSS. The result of entering data into the MDSS is the generation of a list of drug options adjusted for a given patient's medical history, biophysical profile and genetic test results (Figure 1).
Figure 1.
Screen shot of drug options generated by medication decision support system for a 62-year-old patient with depression who is a poor metabolizer of CYP2C19.
Participants
Owing to the known association between HLA-B*58:01 and life-threatening severe cutaneous adverse reactions induced by allopurinol,18 it was not considered ethical to perform a randomized controlled study when including the care of people with gout in a pharmacogenetic study.
We recruited family physicians and pharmacists through their teaching connection with the Department of Family Practice at our institution. Those who showed an interest in pharmacogenomics and had electronic health records were enlisted. The family physicians and pharmacists recruited patients opportunistically or through a search of their electronic health record, including adults at least 18 years of age who were not pregnant or breast-feeding and had a diagnosis of gout, chronic obstructive pulmonary disease, migraine, depression, osteoarthritis, hypertension, hyperlipidemia, atrial fibrillation, osteoporosis and/or epilepsy.
Data collection and DNA isolation, extraction and genotyping
After providing signed, informed consent, patients gave saliva samples using the Oragene•DNA collection kit (DNA Genotek) in the physician's office or pharmacy. The samples were transported to the laboratory by research staff, mail or float plane. We extracted genomic DNA using a magnetic-bead-based extraction method (Ambion MagMAX, Applied Biosystems) (Appendix 1). Each sample was quantified with the Qubit 2.0 fluorometer (Thermo Fisher Scientific), and DNA quality was assessed with the use of the 260-nm and 280-nm absorbance ratio (pure DNA: 1.8-2.0; protein contamination: < 1.8; RNA contamination: > 2.0). We performed single nucleotide polymorphism and copy number variant genotyping by means of the quantitative polymerase chain reaction on the QuantStudio 12K Flex system.
Statistical analysis
The primary outcome was ability to obtain and genotype samples, determined by the number of samples received in the laboratory and the number of genetic reports generated. Samples from all patients who entered the study were evaluated. The secondary outcomes were yield and purity of DNA samples, ability to link results to decision support software and use of the MDSS. All samples were tracked for linkage to the MDSS, which recorded the number of times a physician/pharmacist entered the system and for how many patients they used decision support. We calculated 95% confidence intervals (CIs) for proportions using the Wilson score on OpenEpi version 3.03.
Ethics approval
The University of British Columbia Clinical Research Ethics Board approved the study.
Results
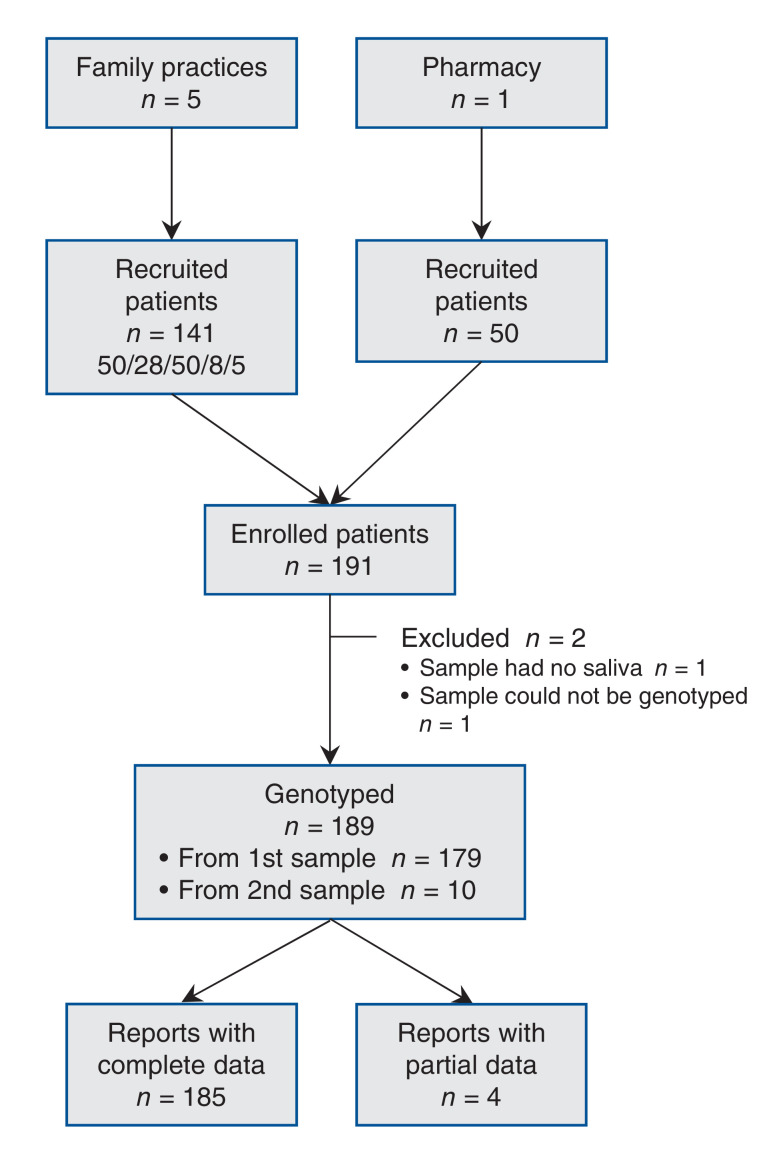
Two family practices on Vancouver Island and 3 family practices and 1 pharmacy in metropolitan Vancouver recruited and obtained saliva samples from a total of 191 patients. The cost of sample collection, extraction and genotyping was $102 per sample. DNA was isolated from 190 samples and was assessed for purity, prepared for analysis and genotyped. The flow through the study is shown in Figure 2.
Figure 2.
Study flow.
Of the 190 saliva samples, 164 (86.3%) were successfully genotyped after the first attempt, 13 (6.8%) were genotyped after the pharmacogenetic panel was rerun, and 1 (0.5%) was genotyped after 3 or more attempts. One sample contained too little DNA. The mean DNA concentration for all attempted extractions (n = 190) was 59.6 ng/µL (95% CI 54.0-65.2 ng/µL). The mean 260/280 absorbance ratio of extracted DNA was 1.87 (95% CI 1.84-1.91).
A second saliva sample was collected from 10 of the 12 patients whose DNA could not be genotyped. This increased the overall success rate to 99.5% (189/190 patients); however, 4 of the recollected samples generated only partial reports.
Of the 185 patients with complete reports with call rates (number of called genotypes per sample divided by total number of genotypes tested) above 95%, 179 (96.8%) had at least 1 actionable genotype for medications included in the MDSS. Single variants were seen in 45 of the 185 patients (24.3%), 65 (35.1%) had 2 variants, and 69 (37.3%) had 3 or more of the variants tested. The complete list of variants and actionable results are found in Table 1. Table 2 shows the actionable results that were found in the 185 participants.
Table 1: Frequency of alleles and diplotypes among patients tested in primary care whose DNA was fully genotyped.
| Gene | Allele/diplotype | No. (%) of patients (n = 185) | 95% confidence interval | Phenotype |
|---|---|---|---|---|
|
SLCO1B1 rs4149056 |
T/T | 132 (71) | 64-77 | Normal function |
| T/C | 49 (26) | 21-33 | Intermediate function | |
| C/C | 4 (2) | 0-4 | Low activity | |
|
G6PD rs5030868 |
C/C | 185 (100) | 98-100 | Normal variant |
| C/T | 0 (0) | 0-2 | Variable activity | |
| T/T | 0 (0) | 0-2 | Low activity | |
|
VKORC1 rs9923231 |
G/G | 38 (20) | 15-27 | Normal activity |
| G/A | 89 (48) | 41-55 | Intermediate activity | |
| A/A | 58 (31) | 25-38 | Low activity | |
| HLA-B*58:01 | *58:01 negative | 176 (95) | 91-97 | Normal |
| *58:01 positive | 7 (4) | 2-8 | Increased hypersensitivity | |
| Undetermined | 2 (1) | 0-4 | Unknown | |
| CYP2C19 | *1/*1 | 80 (43) | 36-50 | Extensive metabolizer |
| *1/*2,*2/*17,*1/*4,*1/*8 | 55 (30) | 24-37 | Intermediate metabolizer | |
| *2/*2,*2/*3,*3/*3 | 8 (4) | 2-8 | Poor metabolizer | |
| *1/*17,*17/*17 | 42 (23) | 17-29 | Ultrarapid metabolizer | |
| CYP2C9 | *1/*1, | 118 (64) | 57-70 | Extensive metabolizer |
| *1/*2,*1/*3,*2/*2,*2/*3 | 62 (34) | 27-41 | Intermediate metabolizer | |
| *3/*3 | 5 (3) | 1-6 | Poor metabolizer | |
|
CYP2D6 Codeine CPIC guidelines† |
*1/*1,*1,*2,*2/*2,*1/*10,(*10/*10)3N,(*2/*4)3N,*1/*17,*1/*3,*1/*4,*1/*5,*1/*41,*2/*10,*2/*4,*2/*5,*2/*9,*2/*41,*10/*10,*41/*41 | 148 (80) | 74-85 | Extensive metabolizer |
| *3/*41,*3/*9,*4/*10,*4/*9,*4/*41,*5/*10,*5/*9 | 19 (10) | 7-15 | Intermediate metabolizer | |
| *3/*4,*4/*14A,*4/*4,*4/*5,*4/*6 | 13 (7) | 4-12 | Poor metabolizer | |
| (*1/*1)xN,(*1/*10)3N,(*1/*2)3N,(*2/*2)3N | 5 (3) | 1-6 | Ultrarapid metabolizer | |
|
CYP2D6 DPWG guidelines† |
*1/*1,*1,*2,*1/*10,*1/*17,*1/*41,*2/*2,*2/*10,*2/*41,*2/*9,(*1/*10)3N | 94 (51) | 44-58 | Extensive metabolizer |
| (*2/*4)3N,*1/*3,*1/*4,*1/*5,*2/*4,*2/*3,*10/*10,(*10/*10)3N, *2/*5,*41/*41,*3/*41,*3/*9,*4/*9,*4/*10,*4/*41,*5/*10,*5/*9 | 74 (40) | 33-47 | Intermediate metabolizer | |
| *3/*4,*4/*14A,*4/*4,*4/*5,*4/*6 | 13 (7) | 4-12 | Poor metabolizer | |
| (*1/*1)xN,(*1/*2)3N,(*2/*2)3N | 4 (2) | 1-5 | Ultrarapid metabolizer |
Note: CPIC = Clinical Pharmacogenetics Implementation Consortium, DPWG = Dutch Pharmacogenetics Working Group.
†Ambiguous genotypes DPWG CYP2D6 (*2/*4)3N and CPIC CYP2D6 (*1/*10)3N were assigned the "worst-case" phenotype.
Table 2: Population with actionable genotypes for drugs within medication decision support system.
| Gene | Drug | No. (%) of participants (n = 185) |
|---|---|---|
| CYP2C19 | Citalopram, escitalopram, esomeprazole, lansoprazole, omeprazole, pantoprazole, sertraline | 50 (27.0) |
| CYP2C19 | Clopidogrel | 63 (34.0) |
| SLCO1B1 | Simvastatin | 53 (28.6) |
| CYP2C9 | Celecoxib, flurbiprofen | 5 (2.7) |
| CYP2C9 | Warfarin | 67 (36.2) |
| VKORC1 | Warfarin | 147 (79.4) |
| G6PD | Sulfamethoxazole | 0 (0) |
| HLA-B*58:01 | Allopurinol | 7 (3.8) |
| CYP2D6 | Metoprolol, oxycodone, propafenone, tramadol, venlafaxine | 91 (49.2) |
| CYP2D6 | Codeine | 18 (9.7) |
Genetic results were linked directly into the MDSS and also were presented to the patient's doctor or pharmacist in a report that included the testing method, drugs implicated and predicted phenotype. The health care professional was able to log in to the MDSS, select his or her patient and then select the relevant condition. Figure 1 shows how, after patient biophysical information is completed, the software displays a list of dosage-adjusted medication options.
The physicians and pharmacists used the software 236 times over 3 months, for 11 of the 15 conditions. The MDSS was most frequently accessed for hyperlipidemia (n = 53) and hypertension (n = 52). It was not accessed for acute gout, migraine, migraine prophylaxis or control of atrial fibrillation rate.
Interpretation
We found that it is feasible to obtain saliva samples in primary care settings and that samples were of sufficient quantity and quality for DNA extraction, purification and genotyping. A total of 97% of patients had at least 1 actionable pharmacogenetic variant. These findings suggest that pharmacogenetic test results can be incorporated into an MDSS. An MDSS that creates medication options adjusted for patient characteristics, drug-drug interactions and pharmacogenetics can be used within primary care.
The challenge of identifying the right medication for the right patient at the right time remains a cascade of failures19 owing in part to the problem of multimorbidity and polypharmacy. Pharmacogenetic testing for variants that are associated with alterations in drug response should help prevent adverse reactions4-6 but adds to the problem of increasing complexity in prescribing. The challenge for health care professionals is how to incorporate this information in the limited time given to deciding on and writing a prescription.20 Preemptive pharmacogenetic testing may be one approach, but the need for a system to help professionals use the genetic information remains.21 It has been suggested that, to affect the level of use of pharmacogenetic results, pharmacists can order pharmacogenetic tests and incorporate results into medication management.22
We have shown that it is possible for both family physicians and pharmacists to obtain saliva samples from patients, send the samples to the laboratory, complete the biophysical and laboratory data needed for an MDSS and make decisions based on individualized medication options. In a pharmacy-based study (n = 54) by Swen and colleagues,23 9 saliva samples (16.7%) contained too little DNA, whereas we had 1 sample (0.5%) that contained too little DNA. We achieved an overall call rate of 99%, compared with Swen and colleagues' call rates of 93.3% and 100% for CYP2D6 and CYP2C19, respectively. Swen and colleagues concluded that pharmacy-initiated pharmacogenetic screening in primary care with respect to quality of DNA collection with saliva kits and genotyping is feasible for a primary care setting; we have exceeded these criteria.
The use of electronic medical records has increased over the last 10 years but has not been associated with a decrease in rates of adverse drug events.2 The use of alerts within the electronic medical record system when selecting a drug is not a successful strategy because such alerts are ignored in 49% to 96% of cases,24 and alerts are unlikely to be effective within the time constraints of usual family practice.25 An alternative to alerts is to build a condition-based MDSS, derived from the clinical guidelines for all conditions, that provides options that are safe and effective for a given patient.15 We built a system that starts with all the possible options for treatment and results in a list of optimal, individualized drug therapy options. These drug options are adjusted for renal and hepatic function, comorbidities, concomitant medications and genetics.
The list of drugs that have associated pharmacogenetic tests includes drugs used in many conditions seen and managed in primary care.26 However, the number of studies involving pharmacogenetic testing in primary care is limited.27 There has been some exploration of clinical decision support including genomics and providing genomic interactions as alerts,28 and the largest study to date showed a significantly lower hospital admission rate among older adults who were tested (9.8%) than among those not tested (16.1%).29 Preliminary results from 2 clinical studies, 1 that recruited from a hospital system and the other from a long-term care facility, showed actionable genotypes for dosage changes or contraindication for the patients' current medications in 24% and 50% of patients, respectively.30,31
Given our finding that 97% of patients had at least 1 actionable pharmacogenetic variant, and the results of a 5000-patient US study in which 96% of participants had actionable pharmacogenetic variants,32 it is likely that future studies will show similar numbers. The high proportion of patients with actionable genotypes, coupled with the fact that 11% of Canadians aged 45-64 years and 30% of those aged 65-79 years take at least 5 prescription drugs concurrently,33 indicates that preemptive administration of a pharmacogenetic test has enormous potential. Pharmacogenetic testing could be of value to preventive medicine: if every patient underwent testing before a medication was needed, when the need arose there would be no need to delay medication or give medication blindly while waiting for a test result.
The development of condition-based prescribing using an evidence-based approach has been discussed.34 We have succeeded in developing and using this approach for multiple drugs and multiple conditions in primary care.
Limitations
This was a small study to assess the feasibility of collecting and analyzing saliva samples and feeding back results to health care professionals within an MDSS. Because we wanted to assess the feasibility of the system and not get involved in other ethical issues around genetic testing, we collected no personal data from patients other than their sex. Consequently, we were unable to present patients' demographic information or discuss patient outcomes. To show the therapeutic utility of an MDSS using pharmacogenetic data, it will be necessary to perform a trial with adequate power and to follow patients to determine outcomes. We will be continuing this work.
Conclusion
Within primary care, it is possible to collect saliva samples of sufficient quantity and quality for DNA extraction, purification and genotyping. These pharmacogenetic tests can be incorporated into a condition-based medication decision support tool that provides a list of dosage-adjusted medication options filtered for potential adverse drug reactions. Further investigation of clinical decision support systems with integrated data from pharmacogenetic tests using patient outcomes is necessary.
Supplemental information
For reviewer comments and the original submission of this manuscript, please see www.cmajopen.ca/content/4/3/E528/suppl/DC1
Appendix
Acknowledgements
The authors acknowledge the work of David J. Chong, Lily Edelson, Nathaniel Heinrichs and Ada Lo in finding and appraising evidence, and developing and refining the logic trees. They thank M. Marta Guarna for her work in the development of the study design and gaining funding. The authors also acknowledge Penelope F. Miller for her time and skill in pharmacy consultation.
Footnotes
Funding: Funding was received from Genome British Columbia, AstraZeneca, GlaxoSmithKline, Health Research Foundation/Rx & D, Janssen Inc., Merck Canada, Pfizer Canada and Hoffmann-La Roche Ltd. The pharmaceutical companies had no influence over the study design, study procedures, formation of decision support, recruitment of participants, data collection, analysis of data, research findings or research publication.
References
- 1.Thomsen LA, Winterstein AG, Søndergaard B, et al. Systematic review of the incidence and characteristics of preventable adverse drug events in ambulatory care. Ann Pharmacother. 2007;41:1411–26. doi: 10.1345/aph.1H658. [DOI] [PubMed] [Google Scholar]
- 2.Taché SV, Sönnichsen A, Ashcroft DM. Prevalence of adverse drug events in ambulatory care: a systematic review. Ann Pharmacother. 2011;45:977–89. doi: 10.1345/aph.1P627. [DOI] [PubMed] [Google Scholar]
- 3.Gandhi TK, Weingart SN, Borus J, et al. Adverse drug events in ambulatory care. N Engl J Med. 2003;348:1556–64. doi: 10.1056/NEJMsa020703. [DOI] [PubMed] [Google Scholar]
- 4.Phillips KA, Veenstra DL, Oren E, et al. Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA. 2001;286:2270–9. doi: 10.1001/jama.286.18.2270. [DOI] [PubMed] [Google Scholar]
- 5.Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte - an update of guidelines. Clin Pharmacol Ther. 2011;89:662–73. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 6.Cargnin S, Jommi C, Canonico PL, et al. Diagnostic accuracy of HLA-B*57:01 screening for the prediction of abacavir hypersensitivity and clinical utility of the test: a meta-analytic review. Pharmacogenomics. 2014;15:963–76. doi: 10.2217/pgs.14.52. [DOI] [PubMed] [Google Scholar]
- 7.Verbeurgt P, Mamiya T, Oesterheld J. How common are drug and gene interactions? Prevalence in a sample of 1143 patients with CYP2C9, CYP2C19 and CYP2D6 genotyping. Pharmacogenomics. 2014;15:655–65. doi: 10.2217/pgs.14.6. [DOI] [PubMed] [Google Scholar]
- 8.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–7. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature. 2015;526:343–50. doi: 10.1038/nature15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cahir C, Fahey T, Teljeur C, et al. Prescriber variation in potentially inappropriate prescribing in older populations in Ireland. BMC Fam Pract. 2014;15:59. doi: 10.1186/1471-2296-15-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stocks SJ, Kontopantelis E, Akbarov A, et al. Examining variations in prescribing safety in UK general practice: cross sectional study using the Clinical Practice Research Datalink. BMJ. 2015;351:h5501. doi: 10.1136/bmj.h5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overhage JM, Gandhi TK, Hope C, et al. Ambulatory computerized prescribing and preventable adverse drug events. J Patient Saf. 2016;12:69–74. doi: 10.1097/PTS.0000000000000194. [DOI] [PubMed] [Google Scholar]
- 13.Weingart SN, Toth M, Sands DZ, et al. Physicians' decisions to override computerized drug alerts in primary care. Arch Intern Med. 2003;163:2625–31. doi: 10.1001/archinte.163.21.2625. [DOI] [PubMed] [Google Scholar]
- 14.Rayo MF, Kowalczyk N, Liston BW, et al. Comparing the effectiveness of alerts and dynamically annotated visualizations (DAVs) in improving clinical decision making. Hum Factors. 2015;57:1002–14. doi: 10.1177/0018720815585666. [DOI] [PubMed] [Google Scholar]
- 15.Kuperman GJ, Bobb A, Payne TH, et al. Medication-related clinical decision support in computerized provider order entry systems: a review. J Am Med Inform Assoc. 2007;14:29–40. doi: 10.1197/jamia.M2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorn CF, Klein TE, Altman RB. Pharmacogenomics and bioinformatics: PharmGKB. Pharmacogenomics. 2010;11:501–5. doi: 10.2217/pgs.10.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Britt H, Miller G. BEACH program update. Aust Fam Physician. 2015;44:411–4. [PubMed] [Google Scholar]
- 18.Ko TM, Tsai CY, Chen SY, et al. Taiwan Allopurinol-SCAR Consortium. Use of HLA-B*58:01 genotyping to prevent allopurinol induced severe cutaneous adverse reactions in Taiwan: national prospective cohort study. BMJ. 2015;351:h4848. doi: 10.1136/bmj.h4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marengoni A, Onder G. Guidelines, polypharmacy, and drug-drug interactions in patients with multimorbidity. BMJ. 2015;350:h1059. doi: 10.1136/bmj.h1059. [DOI] [PubMed] [Google Scholar]
- 20.Slight SP, Howard R, Ghaleb M, et al. The causes of prescribing errors in English general practices: a qualitative study. Br J Gen Pract. 2013;63:e713–20. doi: 10.3399/bjgp13X673739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haga SB, Moaddeb J. Comparison of delivery strategies for pharmacogenetic testing services. Pharmacogenet Genomics. 2014;24:139–45. doi: 10.1097/FPC.0000000000000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bashir NS, Ungar WJ. The 3-I framework: a framework for developing public policies regarding pharmacogenomics (PGx) testing in Canada. Genome. 2015;58:527–40. doi: 10.1139/gen-2015-0100. [DOI] [PubMed] [Google Scholar]
- 23.Swen JJ, van der Straaten T, Wessels JAM, et al. Feasibility of pharmacy-initiated pharmacogenetic screening for CYP2D6 and CYP2C19. Eur J Clin Pharmacol. 2012;68:363–70. doi: 10.1007/s00228-011-1130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Sijs H, Aarts J, Vulto A, et al. Overriding of drug safety alerts in computerized physician order entry. J Am Med Inform Assoc. 2006;13:138–47. doi: 10.1197/jamia.M1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Overby CL, Tarczy-Hornoch P, Hoath JI, et al. Feasibility of incorporating genomic knowledge into electronic medical records for pharmacogenomic clinical decision support. BMC Bioinformatics. 2010;11(Suppl 9):S10. doi: 10.1186/1471-2105-11-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufman AL, Spitz J, Jacobs M, et al. Evidence for clinical implementation of pharmacogenomics in cardiac drugs. Mayo Clin Proc. 2015;90:716–29. doi: 10.1016/j.mayocp.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haga SB, LaPointe NMA, Cho A, et al. Pilot study of pharmacist-assisted delivery of pharmacogenetic testing in a primary care setting. Pharmacogenomics. 2014;15:1677–86. doi: 10.2217/pgs.14.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell GC, Crews KR, Wilkinson MR, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc. 2014;21(e1):e93–9. doi: 10.1136/amiajnl-2013-001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brixner D, Biltaji E, Bress A, et al. The effect of pharmacogenetic profiling with a clinical decision support tool on healthcare resource utilization and estimated costs in the elderly exposed to polypharmacy. J Med Econ. 2016;19:213–28. doi: 10.3111/13696998.2015.1110160. [DOI] [PubMed] [Google Scholar]
- 30.Eadon MT, Desta Z, Levy KD, et al. Implementation of a pharmacogenomics consult service to support the INGENIOUS trial. Clin Pharmacol Ther. 2016;100:63–6. doi: 10.1002/cpt.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saldivar JS, Taylor D, Sugarman EA, et al. Initial assessment of the benefits of implementing pharmacogenetics into the medical management of patients in a long-term care facility. Pharmgenomics Pers Med. 2016;9:1–6. doi: 10.2147/PGPM.S93480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bush WS, Crosslin DR, Owusu-Obeng A, et al. Genetic variation among 82 pharmacogenes: the PGRNseq data from the eMERGE network. Clin Pharmacol Ther. 2016;100:160–9. doi: 10.1002/cpt.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rotermann M, Sanmartin C, Hennessy D, et al. Prescription medication use by Canadians aged 6 to 79. Health Rep. 2014;25:3–9. [PubMed] [Google Scholar]
- 34.Roin BN. Solving the problem of new uses. Rochester (NY): Social Science Research Network; 2013. [accessed 2016 Sept. 7]. Available http://papers.ssrn.com/sol3/papers.cfm?abstract_id=2337821.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.