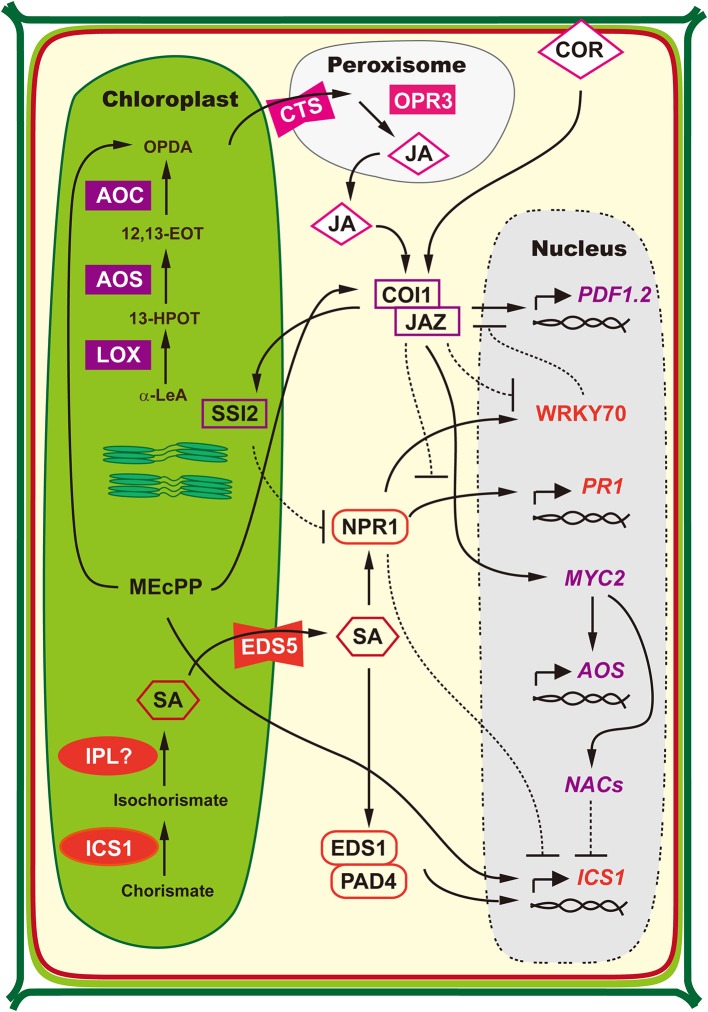
Figure 2.
Regulation of SA and JA Biosynthesis is Associated with Chloroplast. SA biosynthesis is predominantly accomplished by nucleus-encoded chloroplast-located isochorismate synthase (ICS1). In chloroplasts, ICS catalyzes the conversion of chorismate into isochorismate, which is further converted to SA by undetermined isochorismate pyruvate lyase (IPL). The MATE-transporter ENHANCED DISEASE SUSCEPTIBILITY 5 (EDS5) is responsible for SA transportation from chloroplast into cytosol. Defense-elicited ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) and PHYTOALEXIN DEFICIENT 4 (PAD4) complex works in a positive feedback loop to control SA synthesis, which is regulated by SA. While in a negative feedback loop, accumulation of ICS1-produced SA results in the deoligomerization of NON-EXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1), which is then translocated into nucleus where it suppresses the ICS1 expression (modified from Boatwright and Pajerowska-Mukhtar, 2013; Seyfferth and Tsuda, 2014). JA biosynthesis originates from polyunsaturated fatty acids released from chloroplast membranes. Firstly, α-linolenic acid (18:3) (α-LeA) is catalyzed by lipoxygenase (LOX) to yield the 13-hydroperoxy derivative 13(S)-hydroperoxy-octadecatrienoic acid (13-HPOT). The dehydration of 13-HPOT by allene oxide synthase (AOS) results in the formation of unstable 12, 13(S)-epoxy-octadecatrienoic acid (12,13-EOT), which is the committed step of JA biosynthesis. Then the 12,13-EOT is converted to 12-oxophytodienoic acid (OPDA) by allene oxide cyclase (AOC) through cyclization and concludes the chloroplast-localized part of JA biosynthesis. Subsequently, OPDA is released from chloroplasts and taken up into peroxisomes by transporter COMATOSE (CTS3). The remaining steps are located in peroxisomes and JA is generated through reduction of the cyclopentenone by OPDA reductase 3 (OPR3) and subsequent three cycles of β-oxidation for side-chain shortening. The JA co-receptor complex of CORONATINE INSENSITIVE1 (COI1) and the negative regulator JAZMONATE ZIM DOMAIN (JAZ) proteins regulates the positive feedback loop of JA biosynthesis. Formation of JA subjects JAZ to proteasomal degradation, which allows MYC2 to activate the JA biosynthesis genes such as AOS, AOC, and LOX (modified from Wasternack, 2007; Schaller and Stintzi, 2009; Wasternack and Hause, 2013). NPR1 is the central transcriptional regulator of SA-mediated defense responses and directly regulates PATHOGENESIS-RELATED 1 (PR1) expression (Wang et al., 2006). By wounding or JA treatment, COI1–JAZ co-receptor promotes the degradation of JAZ and release the positively acting transcription factors that binds to JA-responsive promoters to initiate the transcription of JA-responsive genes, such as PLANT DEFENSIN1.2 (PDF1.2) (Chini et al., 2007; Thines et al., 2007; Yan et al., 2009). During the antagonistic interplay between SA and JA, NPR1 suppresses COI1-JAZ mediated induction of JA-responsive genes via WRKY transcription factors, while JA also represses WRKY in COI1-dependent pathway (Li et al., 2004; Gao et al., 2011). On the other hand, the JA signaling proteins, such as chloroplast factor SUPPRESSOR OF SA INSENSITIVITY 2 (SSI2), negatively regulate SA-mediated NPR1-dependent defense responses (Kunkel and Brooks, 2002). Further, the phytotoxin coronatine (COR), a molecular mimic of JA, activates NAC transcription factors via COI1-JAZ and MYC2, which eventually inhibits SA accumulation through repressing ICS1 expression (Zheng et al., 2012). In addition, the stress-induced methylerythritol cyclodiphosphate (MEcPP) acts as a plastid-to-nucleus retrograde signal to increase the transcription level of ICS1 (Xiao et al., 2012). Meanwhile, MEcPP increase the level of JA precursor OPDA and induce JA-responsive genes via a COI1-dependent manner in the presence of high SA (Lemos et al., 2016). Solid lines with arrow head represent activation or promotion, dotted lines with bar head to represent deactivation or inhibition.