FIGURE 1.
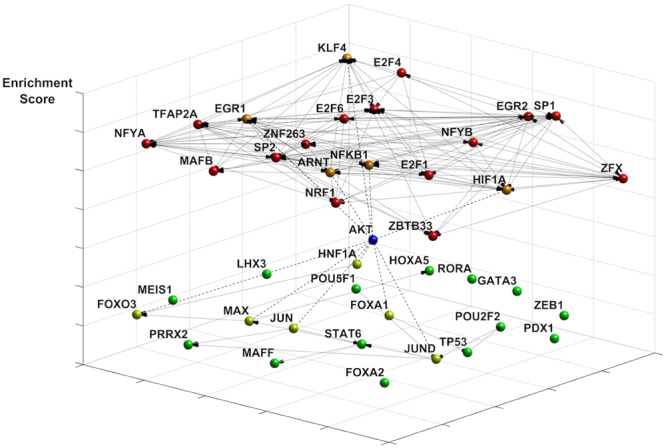
Effects of ATP restriction on gene regulatory networks in quiescent fibroblasts. The transcription factor (TF) network was constructed using the most enriched TFs motifs (JASPAR 2014 database). Co-regulatory connections for these motifs are provided by DNaseI footprinting (see text). Represented are the most enriched (p < 0.005, coded in red) and avoided (p > 0.998, coded in green) motifs, whereby p-values indicate the likelihood for a motif to occur against the background of 263 promoters in all known genes. Enriched motifs indicate a highly connected network (20 nodes, 127 edges), with many highly connected nodes including the NF-κB TF binding motif NFKB1. The network of avoided TF motifs is only sparsely connected (20 nodes, 16 links). The differentiation of the motifs by energy restriction predicts a role of Akt signaling in shaping changes in gene expression. Akt connects to TFs in the enriched group (dotted line, orange), such as EGR1, KLF4, and NFKB1. TFs known to be inhibited by Akt include FOXO TFs, HNF1A, and JUN (dotted line, neon green).
