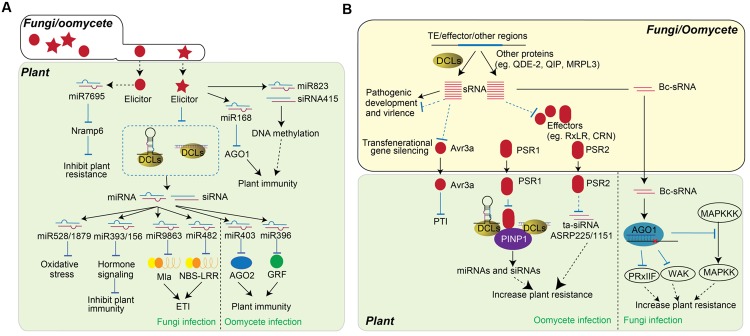
FIGURE 2.
Role of sRNAs in plant-fungi/oomycete interaction. (A) Plant sRNAs regulate PTI and ETI in response to fungi or oomycete infection. The infection of fungi (left) and oomycete (right) alters the accumulation of miRNAs, by which changes the expression of genes contribute to plant resistance. Fungi elicitorsor fungi infections triggers the accumulation of some sRNAs, such as miR7695, miR168, miR823 and siRNA415, while miR528, miR1879, miR9863, and miR482 are down-regulated to improve plant resistance. The accumulations of miR403 and miR396 are down-regulated upon oomycete infection. (B) Schematic representation of the function of fungi/oomycete sRNAs in pathogen virulence. sRNAs encoded by fungi and oomycetes are usually generated from TE region, effector coding region, and other regions. These sRNA can be either DCL-dependent or DCL-independent and are involved in the regulation of pathogen development and virulence. In particular, sRNA regulate the expression of effectors, which further influence the accumulation of host miRNA and siRNA. sRNAs generated from Avr3a region of oomycete can transgenerationally change the pathogen virulence. The PSR1 and PSR2 effectors of oomycete are secreted into plant cells and alter host RNA silencing machineries as RNA silencing suppressor to decrease host immunity. On the other hand, fungi sRNAs, Bc-sRNAs, translocate into host cell and utilize plant RNA silencing component to reduce the expression of host immune genes and facilitate fungi infection.