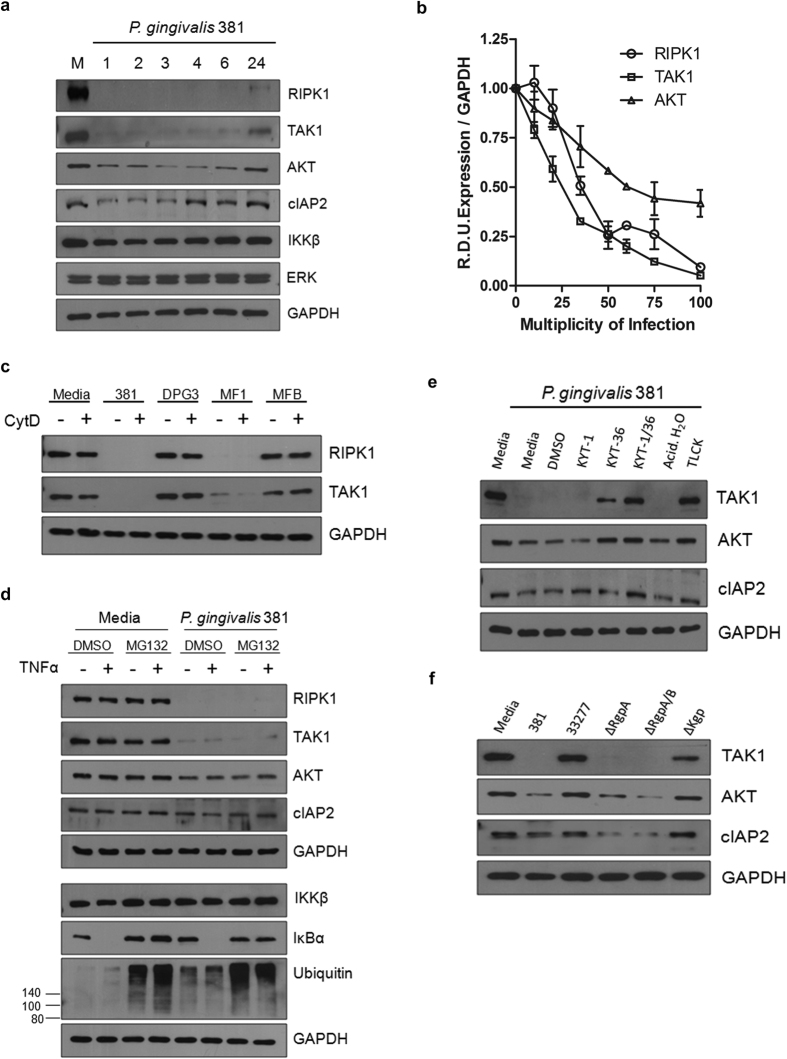
Figure 2. P. gingivalis degrades TAK1 and AKT and proteolysis is dependent on Kgp activity and major fimbriae.
(a) HUVEC were left untreated (M: media) or cocultured with 381 MOI 100 for 1, 2, 3, 4, 6, or 24 hr. (b) HUVEC were cocultured with 381 MOI 10, 20, 35, 50, 65, 75, or 100 for 2 hr. Densitometry was performed on three independent membranes to calculate RIPK1 R.D.U. relative to GAPDH loading control. (c) P. gingivalis 381 or fimbriae mutant strains (major fimbriae mutant -DPG3, minor fimbriae mutant -MF1, or major and minor fimbriae mutant -MFB) were pretreated with DMSO or cytochalasin D (1 μg/mL) for 30 min and then immediately cocultured with HUVEC at an MOI 100 for 2 hr. (d) HUVEC were pretreated with vehicle control (DMSO) or MG132 for 2.5 hr (30 μM) and then left untreated or cocultured with P. gingivalis 381 for 2 hr, followed by restimulation with 10 ng/mL TNFα for 15 min. (e) P. gingivalis was pretreated with selective gingipain inhibitors (Rgp-specific: 10 μM KYT-1, Kgp-specific: 10 μM KYT-36, 10 μM KYT-1 and 10 μM KYT-36), the general cysteine inhibitor TLCK (1 mM), or vehicle controls for 45 min. HUVEC were then immediately cocultured with medium or pretreated preparations of P. gingivalis for 2 hr. (f) HUVEC were cocultured with wild-type strains 381, 33277, or gingipain mutant strains (ΔRgpA, ΔRgpA/B, ΔKgp) at an MOI 100 for 2 hr. (a–f) Whole cell lysates were analyzed for RIPK1, TAK1, cIAP2, IKKβ, P- IKKβ, IκBα, P-IκBα, AKT, ubiquitin, ERK, or GAPDH via Western blot analysis.