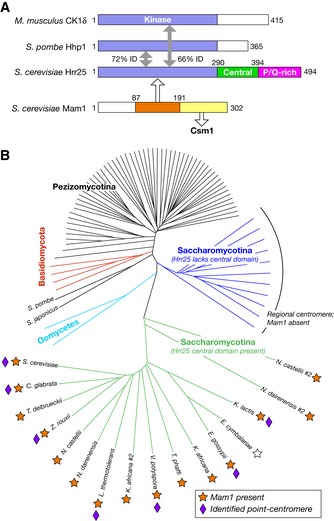
Phylogenetic tree constructed from an alignment of 93 fungal Hrr25 orthologs. Branches in green represent Hrr25 proteins from Saccharomycotina yeast with the conserved central domain (see Fig
EV1A for sequence alignment). Branches in blue represent Saccharomycotina Hrr25 proteins lacking this domain. Orange stars indicate the presence of a Mam1 ortholog in each species' genome.
E. cymbalariae (white star) possesses an unannotated Mam1 ortholog on chromosome VII (region ˜313,000–315,000, 32% identity to
S. cerevisiae Mam1 residues 83–242, identified by TBLASTN (NCBI)). Purple diamonds denote organisms in which point‐centromere sequences have been identified (Meraldi
et al,
2006; Gordon
et al,
2011). Point centromeres and Mam1 orthologs are both exclusive to the group of yeast whose Hrr25 orthologs possess the central domain.