Figure 2. Structure and nucleotide/inhibitor binding by budding yeast Hrr25.
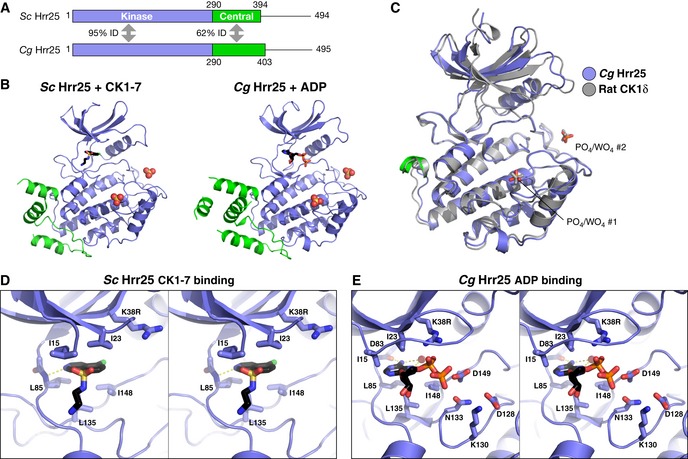
- Domain diagram of crystallized Hrr25 constructs from Saccharomyces cerevisiae and Candida glabrata, missing the P/Q‐rich C‐terminal domain.
- Structures of S. cerevisiae Hrr251–394 K38R bound to CK1‐7 (Chijiwa et al, 1989; Xu et al, 1996) and C. glabrata Hrr251–403 K38R bound to ADP, with domains colored as in (A). Bound / ions are shown as spheres (see Fig EV2A for additional C. glabrata Hrr25 structures and Fig EV3A for ion electron density).
- Stereo view of CK1‐7 binding to S. cerevisiae Hrr25. Bound drug is positioned identically to a previous structure of S. pombe Cki1 bound to CK1‐7 (Xu et al, 1996). All active‐site residues shown are conserved between S. cerevisiae and C. glabrata Hrr25.
- Stereo view of ADP binding to C. glabrata Hrr25 (formate structure; structure is equivalent, but the GxGxxG motif is disordered in that structure). All active‐site residues shown are conserved between S. cerevisiae and Candida glabrata Hrr25.
