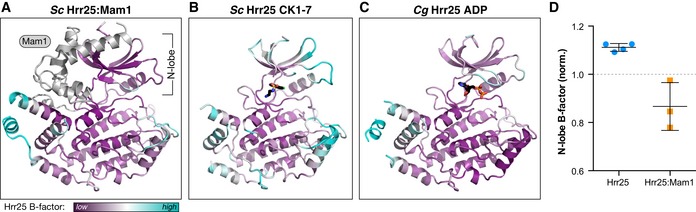
Figure 6. Mam1 binding restricts Hrr25 N‐lobe mobility.
-
A–CRibbon views of the Saccharomyces cerevisiae Hrr251–394:Mam187–191 structure (form 1), CK1‐7 bound S. cerevisiae Hrr251–394, and ADP‐bound C. glabrata Hrr251–403 (formate condition), with Hrr25 colored according to main‐chain B‐factor from low (purple) to cyan (high). For each, coloring is normalized to correspond to the average of the 20 lowest (purple) or highest (cyan) main‐chain B‐factors in Hrr25. Mam1 (panel A) is colored in light gray.
-
DOverall main‐chain B‐factors for the N‐lobe (residues 1–85) of four structures of Hrr25 (both C. glabrata and S. cerevisiae) and the three crystallographically unique views of the S. cerevisiae Hrr25:Mam1 complex. Values are normalized to the average main‐chain B‐factor for the entire Hrr25 chain (dotted line at 1.0). P‐value = 0.004 (Student's t‐test) (see Fig EV5 for residue‐by‐residue B‐factor plots for each structure). Error bars represent standard deviation.
