Figure 5. LincGET acts as both a transcription factor and an exon skipping splicing inhibitor.
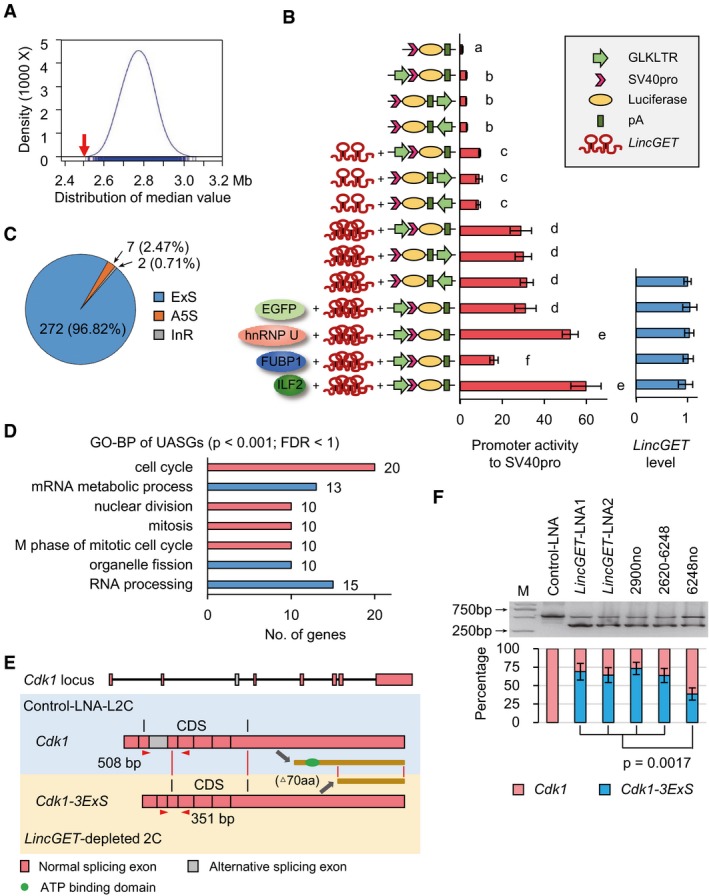
- The median distance of DEGs to neighbor GLKLTRs (red arrow) and the distribution of that of random genes (blue) (P < 2.2 × 10−16) measured by Wilcoxon rank single test. GLKLTRs are gene loci that contain whole LTR of GLN, MERVL, or ERVK. The results show that DEGs prefer neighboring to GLKLTRs.
- Dox‐induced LincGET expression assay and dual‐luciferase reporter system show that LincGET increased the enhancer activity of GLKLTRs in 293T cells in a dose‐dependent manner. The enhancer activity of GLKLTRs is also increased by overexpression of hnRNP U or ILF2 and decreased by FUBP1 overexpression, which have no effect on LincGET levels. The y‐axis shows the construction of luciferase reporter plasmids and overexpressed genes. Three experimental replicates were performed. Two‐tailed Student's t‐test was used for the statistical analysis. Different letters indicate significant difference (P < 0.01).
- Number and percentage of unusual alternative splicing genes (UASGs, FDR < 0.05) in LincGET‐depleted 2C compared to control‐LNA L2C. ExS, exon skipping; A5S, alternative 5′ splicing; InR, intron retention.
- GO‐BP analysis of UASGs showed that “cell cycle” especially the “M phase of the mitotic cell cycle” is primarily affected. The terms with P < 0.001 and FDR < 1 are shown.
- Cdk1 was exon skipping spliced in LincGET‐depleted 2C, resulting in Cdk1‐3ExS where the third exon was deleted. The abnormal spliced Cdk1‐3ExS variant translated into a truncated CDK1 that lacks the N‐terminal 70 amino acids (aa), which affects the kinase activity of CDK1 lacking a conservative ATP binding domain predicted by InterPro. PCR primers for panel (F) (Cdk1‐Ex3‐F and Cdk1‐Ex3‐R, sequences are shown in Table EV1) are shown in red triangles.
- Exon skipping event of Cdk1‐3ExS in LincGET‐depleted 2C is confirmed by RT–PCR. Injection of LincGET fragments at the pronuclear stage was unable to inhibit the exon 3 skipping of Cdk1, while injection of full‐length LincGET1 lacking LincGET‐LNA2 target site partially inhibited the exon 3 skipping of Cdk1. For each lane, about 50 embryos were used. The error bars represent s.e.m. Three experimental replicates were performed. Two‐tailed Student's t‐test was used for the statistical analysis.
