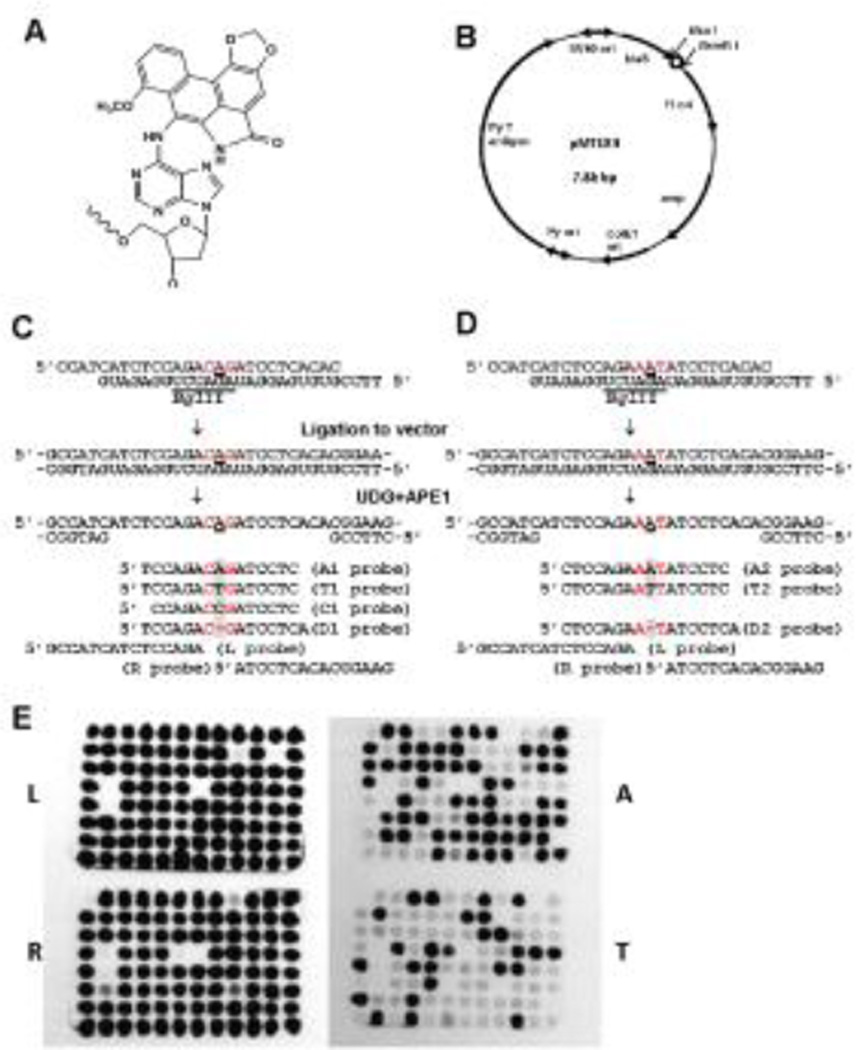
Fig. 1. Preparation of dA-AL-I-bearing plasmid and oligonucleotide probes.
(A) Chemical structure of dA-AL-I; (B) Structure of a MEF–E. coli shuttle vector: Py, mouse polyoma virus; ori, replication origin; amp, ampicillin resistance gene; blaS, blasticidin S-resistance gene; the dA-AL-I insertion site (open circle) is located between BsaI and BsmBI.; (C) and (D) Preparation of two gapped plasmids and probes employed: they differ only in the immediate neighboring bases (shown in red) flanking the adduct. A represents dA-AL-I. Note three mismatches at 5’CAG/5’AGA and two mismatches at 5’AA/5’GA. A1, A2, T1, T2, C1, D1, and D2 probes detect a coding event at dA-AL-I, and L and R probes confirm the presence of the inserted modified oligonucleotide; and (E) Examples of colony hybridization with oligonucleotide probes. Colonies showing positive hybridization signals of both L and R probes are considered to be derived from TLS. E. coli transformants that did not hybridize to L and R probe were excluded from analysis. Probes A and T detect A→A correct TLS events and A→T transversions, respectively.