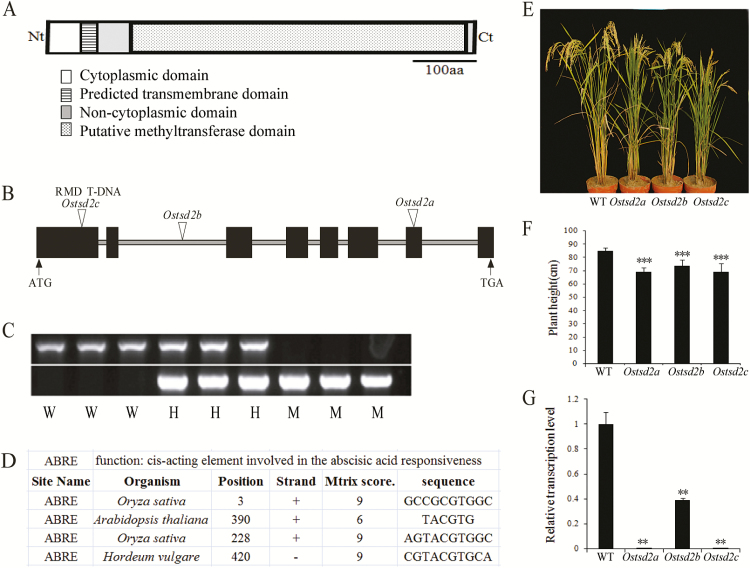
Fig. 1.
OsTSD2 protein structure, gene structure, promoter function prediction, identification, and morphological phenotype of Ostsd2 mutants. (A) OsTSD2 protein structure. A transmembrane domain is predicted at the N-terminal (Nt). A putative methyltransferase domain is predicted at the C-terminal (Ct). (B) OsTSD2 gene structure and T-DNA insertion lines of mutants. Exons are shown as black boxes and introns are shown as lines. The initiator ATG and stop codon TGA are shown. Triangles indicate the insertion sites of three T-DNA lines. (C) Identification of Ostsd2 mutants by PCR. W, wildtype; H, heterozygous; M, homozygous. (D) Four ABREs were found in OsTSD2 using the promoter functional prediction tool (PlantCARE). (E) Morphological phenotype of three Ostsd2 mutants compared with the wildtype (WT, ZH11) at the mature stage. (F) Plant height of WT and three Ostsd2 mutants. (G) OsTSD2 transcription levels in roots of WT and three mutants. Results of Student’s t-test: *, P<0.05; **, P<0.01; ***, P<0.001. (This figure is available in color at JXB online.)