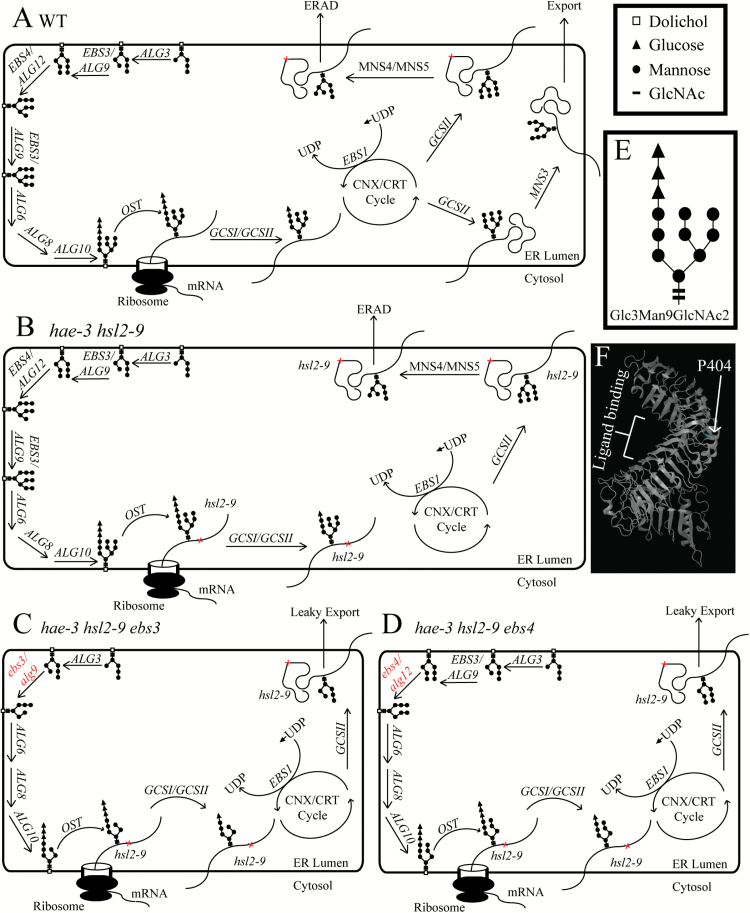
Fig. 6.
Hypothetical model of N-glycan assembly and protein folding. (A) Wild-type cells sequentially add mannose and glucose molecules to assemble the N-glycan Glc3Man9GlcNAc2 in the ER. Glycans are then transferred onto nascent polypeptides before entering the CNX/CRT cycle of protein folding. Correctly folded proteins are eventually exported, while terminally misfolded proteins are sent for degradation (both of these paths are dependent upon trimming N-glycans to expose specific mannose residues). (B) The hae-3 hsl2-9 mutant correctly assembles Glc3Man9GlcNAc2, and polypeptides enter the protein folding cycle, but structural imperfections in hsl2-9 prevent correct folding (note that hae-3 is also likely to be misfolded as well). These proteins will be sent for degradation. (C, D) The hae-3 hsl2-9 ebs3 (C) and hae-3 hsl2-9 ebs4 (D) mutants do not assemble complete N-glycans, preventing α1,6-linked mannose residues from being exposed to signal receptors for degradation. This allows mutant hsl2-9 receptor to leak out of the ER and be trafficked to the plasma membrane to signal to regain abscission. Note that the ebs3 mutation prevents downstream action of EBS4, and assembly of the glycan is halted (C). Similarly, the ebs4 mutation prevents EBS3 from adding a second mannose residue, also halting glycan assembly (D). (E) Structure of Glc3Man9GlcNAc2. (F) Theoretical structure of the HSL2 receptor showing the P404 residue and putative ligand-binding region. (This figure is available in colour at JXB online.)