Abstract
Extracellular pH has a strong effect on cell metabolism and growth. Precisely detecting extracellular pH with high throughput is critical for cell metabolism research and fermentation applications. In this research, a series of ratiometric fluorescent pH sensitive polymers are developed and the ps-pH-neutral is characterized as the best one for exculsive detection of extracellular pH. Poly(N-(2-hydroxypropyl)methacrylamide) (PHPMA) is used as the host polymer to increase the water solubility of the pH sensitive polymer without introducing cell toxicity. The fluorescent emission spectra from the polymeric sensor under excitation at the isosbestic point 455 nm possess two fluorescence peaks at 475 nm and 505 nm, which have different responding trends to pH. This enables the polymer to detect pH using fluorescent maxima at 475 nm and 505 nm (I475nm/I505nm) ratiometrically. The cell impermeability ensures the sensor can solely detect the environmental pH. The sensor is tested to detect the extracellular pH of bacteria or eukaryotic cells in high throughput assays using a microplate reader. Results showed that the pH sensor can be used for high throughput detection of extracellular pH with high repeatability and low photobleaching effect.
Graphical Abstract
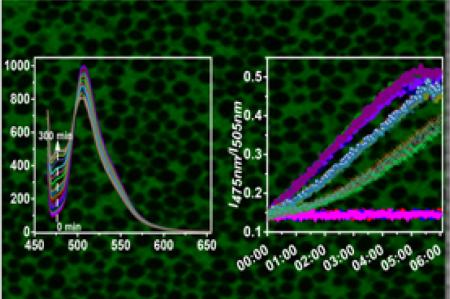
A polymeric water-soluble extracellular pH sensor is developed with fluorescence ratio-metric and cell membrane impermeable characters. The pH sensor enables us to exclusively detect the environmental pH of cells in real time, including bacteria, yeast and mammalian cells. The capability to detect extracellular pH under multi-conditions is also achieved by applying this pH sensor to cell cultures in microplates and detecting fluorescence changes with microplate-reader.
1. Introduction
Although the intracellular pH of microbes and mammalian cells are precisely regulated by homeostasis for the maintenance of metabolism and cell growth, the extracellular pH can vary greatly 1-5. Due to a reprogrammed metabolic pathway and changes of nutrition and oxygen supply, cells commonly exocytose protons or acid metabolites, which results in the acidification of the extracellular microenvironment. Typically, this includes microbes in fermentation and cancer cells 6-10. Environmental pH has many effects on cells’ behavior. It is well known that an acidic extracellular microenvironment drives tumor cell invasion and metastasis, affects microenvironmental immune function, and influences non-cancerous cell growth and differentiation 11-16. The extracellular pH of cancer cells is an important parameter that reflects cell growth and responses to treatment. Extracellular pH also affects the growth and production of microbe metabolites. It is a critical parameter in microbial fermentation which needs to be dynamically monitored and well controlled 16-23. Even though an electrode pH meter can detect the extracellular pH in a large volume, it has shortcomings in analyzing multiple samples with high throughput. A pH sensor with the ability to efficiently and accurately monitor extracellular pH is highly desired for both industry and biomedical research.
Our group has developed a series of cell metabolism sensors, including a series of fluorescence pH sensors 24-28. Because of the high sensitivity, simplicity and the feasibility of miniaturization for microenvironmental detection, fluorescence pH sensors attract significant attention and have been experiencing a series of successes, most of which target the cytosol or some organelles, i.e. mitochondria and lysosomes 28-35. Only a few of them can be specifically limited outside of a cell by either fixing a sensing film on a substrate or anchoring the sensor complex onto the cell surface 27, 28, 36. There is no doubt that monitoring the pH of the cytosol or specific organelles plays an important role in understanding cellular responses to different stresses. However, detection of pH changes that occur in the extracellular microenvironment of cells is also pivotal to understanding the physiological response of cancer cells during treatment or tumor development 37, 38, as well as to the optimization of microbial metabolism and fermentation 39, 40.
While the sensitivity of fluorescence pH sensors is advantageous, the fluorescence intensity is very easily affected by excitation intensity and the distribution of sensors, which results in a low accuracy of pH measurement. To reduce this negative effect, users must make a standard pH titration curve with the same conditions when they used the sensor to detect the pH. Recently, ratiometric detection has proven be a more accurate method by introducing another emission peak into the sensors 29, 41-45. Ratiometric sensors normally possess two emission peaks which have different responses to the same pH. The only factor affecting the ratio of emission intensity is pH; neither the local concentration of sensors nor the exciting intensities change the ratios between peaks in a suitable range.
Stimulated by the research on ratiometric pH sensing probes, we developed a water soluble, pH sensing polymer with undetectable cell toxicity41. Unlike the small molecular sensing probe, this polymeric sensor is exclusively extracellularly localized with the ability to ratiometrically detect pH with high accuracy. By simply mixing and measuring, our research shows that it will enable researchers to perform high-throughput measurements of extracellular pH with a commercial microplate reader, which is commonly used in cell-based high-throughput assays 46-49.
2. Materials and methods
2.1 Materials and reagents
All chemicals and reaction solvents were of analytical grade and were used without further purification. N-(2-hydroxyethyl)-4-bromine-1,8-naphthalimide, 3-amino-1,2,4-triazole 6-bromohexanoic acid , dichloromethane, methanol, triethylamine, methacryloyl chloride, N, N’-dimethylformamide (DMF), dimethyl sulfoxide (DMSO), -2-propanol, [2-(methacryloyloxy)ethyl]trimethylammonium chloride (MAETMA), 4-dimethylaminopyridine (DMAP), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), and azobisisobutyronitrile (AIBN) were commercially available from Sigma-Aldrich (St. Louis, MO) and used without further purification. 2-(Methacryloyloxy)ethylsulfonic acid sodium salt (MAESA) was purchased from Fisher Scientific. N-(2-hydroxypropyl)methacrylamide (HPMA) was synthesized and purified according to a published method 25, 50. The pH sensing probe ANNA, which was named by Zhou J. et al (Scheme 1), was synthesized according to a modified procedure in the literature 41. Deionized water was used for the preparation of buffer solutions. The pH values were determined with a digital pH meter (Thermo Electron Corporation, Beverly, MA) and calibrated at room temperature with standard buffers.
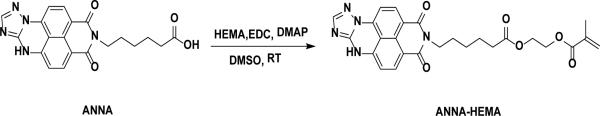
Scheme 1.
Synthesis of the monomeric pH sensing probe (ANNA-HEMA)
2.2 Instruments
A Varian liquid-state NMR operated at 400 MHz for 1H NMR was used for NMR spectra measurements. A Shimadzu UV-3600 UV-Vis-NIR spectrophotometer (Shimadzu Scientific Instruments, Columbia, MD) was used for absorbance measurements. A Shimadzu RF-5301 spectrofluorophotometer was used for fluorescence measurements. Waters Breeze gel filtration chromatography (GPC) was used for polymer molecular weight measurement. Synergy™ H4 Hybrid Multi-Mode Microplate Reader (BioTeK) was used for pH measurement with standard 96-well plates. A Beckman DU 530 UV/Vis Spectrophotometer (Beckman Coulter) was used to measure the microbe density (OD600nm). A Confocal microscope (Nikon, TE2000E) was used for cell imaging.
2.3 Synthesis of the monomeric of pH probe (ANNA-HEMA)
The polymerizable pH probe, ANNA-HEMA, was synthesized according to Scheme 1. 100 mg (0.26 mmol) of ANNA, EDC (121 mg, 0.78 mmol) and DMAP (93.5 mg, 0.78 mmol) were dissolved in 3 mL of DMSO. 102 mg of HEMA (0.78 mmol) was slowly added into the above mixture at room temperature. The reaction was stirred overnight at room temperature. 150 mL of dichloromethane was added to the reaction mixture. The organic phase was washed with ice-cold water twice and one time with brine and then dried over MgSO4. The product of ANNA-HEMA was purified by silica column chromatography with methylene chloride/methanol (95:5 by volume) containing 0.3% triethylamine. Yield: 100 mg (78.1%). 1H NMR (CDCl3, ppm, δ,): 8.61 (d, 1H), 8.40 (d, 1H), 8.04 (d, 1H), 7.52 (d, 1H), 6.88 (d, 1H), 6.10 (s, 1H), 5.57 (s, 1H), 4.31 (s, 4H), 4.18 (t, 2H), 2.35 (t, 2H), 1.72 (m, 1.76~1.68, 4H), 1.46 (m, 1.50~1.42, 2H). MOLDI-TOF (m/z): 504.19, calcd: C26H25N5O6, (M+H), 504.18.
2.4 Polymerization and characterization of the pH sensors (ps-pH)
250 mg of HPMA, 6.0 mg of ANNA-HEMA, 30 mg of MAETMA or MESA (for ps-pH-positive or ps-pH-negative, respectively), and 5.0 mg AIBN were dissolved in 3 mL of DMF. This solution was degassed three times through a standard freeze-thaw process. The monomers were polymerized at 65 °C for 16h under nitrogen. The polymer was precipitated into 150 mL of acetone from the DMF solution. The polymer was re-dissolved in 3 mL methanol and re-precipitated into 100 mL of ether. This produced 198 mg of ps-pH-neutral (yield: 77.3%), 190 mg of ps-pH-negative (yield: 66.4%), 176 mg of ps-pH-positive (yield: 61.5%). The sensor's contents in polymers, which were determined by UV absorbance at pH 7.0, are all around 0.02 g per gram of polymers.
2.5 Culture of L. fermentum (Lactobacillus fermentum) for cellular distribution assay and extracellular pH sensing
L. fermentum (ATCC® 9338™) were cultured in lactobacilli de Man-Rogosa-Sharpe (MRS) broth following the culture method provided by the supplier. The concentration of lactobacilli was estimated by measuring the optical density at 600 nm (OD600nm). According to the amount of cells designated for experiments, an appropriate volume of culture was spun down to harvest cells. The final pellet was re-suspended into fresh MRS medium with or without 10 μg/mL of ps-pH sensor to get the required concentration for experiments. Final concentration of the pH probe in the analysis solution is 0.4 μM.
2.6 Culture of E. coli (Escherichia coli) and B. subtilis (Bacillus subtilis) for cellular distribution assay and extracellular pH sensing
E. coli (JM109) or B. subtilis (168) were cultured in Luria-Bertani (LB) broth overnight at 37°C with vigorous shaking at 180 rpm. The concentrations of bacteria in culture were estimated by measuring the optical density at 600 nm (OD600nm). Bacteria were harvested from the appropriate volume of culture by spin-down according to the amount of cells expected for experiments. The final pellet was re-suspended into fresh LB medium with or without 10 μg/mL of ps-pH sensor to get the required concentration for experiments.
2.7 Culture of S. cerevisiae (Saccharomyces cerevisiae) for cellular distribution assay and extracellular pH sensing
S. cerevisiae (ATCC® 9763™) were cultured in yeast extract peptone dextrose (YEPD) medium overnight at 30 °C with vigorous shaking at 180 rpm. The culture was diluted with fresh YEPD medium, followed by additional two hours of incubation at 30°C. According to the concentrations of yeast in culture which was estimated by measuring the optical density at 600 nm (OD600nm), yeast were harvested from the appropriate volume of culture by spin-down. The final pellet was re-suspended into fresh YEPD medium with or without 10 μg/mL of ps-pH sensor to get the required concentration for experiments.
2.8 Culture of HeLa cells and MCC-7 cells for cellular distribution assay and extracellular pH test
Both HeLa and MCF-7 cell lines were purchased from ATCC. Cells were seeded in a standard 96-well plate and cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin and incubated at 37 °C in a 5% CO2 atmosphere. After getting 100% of confluence, each well was washed by PBS once and the fresh DMEM adjusted to different pH (medium-pH) was added to each well, respectively. The Medium-pH containing 10 μg/mL sensors (ps-pH-neutral) and medium only were used as control in the experiment.
2.9 Cellular distribution of pH sensors assay by fluorescent microscopy
Mammalian cells or microbes were seeded into a 96-well plate and incubated in their medium containing 10 μg/mL pH sensors for 24 h. For mammalian cells (HeLa cell, MCF-7 cell or J774 cell), MitoTracker® Red FM (ThermoFisher Scientific) were added into medium and incubated for 4h before imaging. The localization of pH sensors were detected by co-focal fluorescence microscopy under 488 nm excitation.
2.10 Detect the microbial growth and pH change of culture
Around 0.1 OD600 of fresh microbial culture containing 10 μg/mL of pH sensors (ps-pH-neutral) was sealed into a cuvette and incubated in a water bath at an optimal temperature specific to them. After re-suspending cells by up-down shaking, cell density was measured by OD600. The spectra of pH sensors in culture were detected by a spectrofluorophotometer under the condition of 455 nm excitation. A pH value was calculated based on ratios of emission intensity at 475 nm (I475nm) to emission intensity at 505 nm (I505nm).
2.11 Detect the extracellular pH with a microplate reader
Microbes were re-suspended into fresh medium containing 10 μg/mL ps-pH-neutral sensor and aliquot into 96-well plate by 100 μl per well. Medium containing either sensor or microbes only were aliquoted into parallel wells as experimental controls. After sealing wells with mineral oil to prevent oxygen exchange, the emission intensities at 475 nm and 505 nm from each well were immediately monitored by a microplate reader with 455 nm of excitation.
3. Results and discussion
3.1 Design and synthesis of polymeric pH sensors
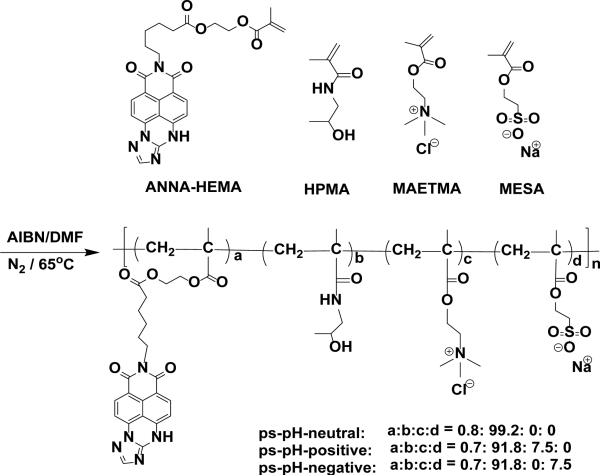
The monomer of the pH sensing probe was synthesized according to Scheme 1. Three kinds of polymeric pH sensors carrying different charges were synthesized according to the scheme in Scheme 2. The molar ratio of monomers used in reaction (a: b: c or d) was 0.7: 91.8: 7.5. We use poly(N-(2-hydroxypropyl)methacrylamide) (PHPMA), which has been widely used as the biocompatible host polymer, to increase the water solubility of sensors 25, 51, 52. A small fraction of poly[2-(methacryloyloxy)ethyl]trimethylammonium chloride (PMAETMA) or poly[2-(methacryloyloxy)ethyl]sulfonic Acid Sodium (PMAESA) was introduced into the polymers to get sensors with positive charges (ps-pH-positive) or negative charges (ps-pH-negative), while the neutral sensor (ps-pH-neutral) does not possess either of these fractions.
Scheme 2.
Chemical structures of the monomers used for preparing the sensing polymers and a polymerization scheme.
The sensors were polymerized using the traditional radical polymerization approach with co-polymerization of ANNA-HEMA, HPMA and MAETMA or MAESA in DMF where AIBN was used as an initiator. The polymeric pH sensors (ps-pH) were sequentially harvested by precipitation from solvent into acetone and ether. To remove any non-polymerized monomers and other potential chemicals, sensors were dialyzed against deionized water for 48h before further characterization and application. The sensors were characterized using gel permeation chromatography (GPC). The average molecular weights (Mn) of three kinds of pH sensors were about 4,000 each. The polydispersity indexes (PDI) of three polymers were about 1.5 each. Zeta potential measurement was performed in 10 mM of HEPES buffer (pH 7.4) and indicated 10.1 mV for ps-pH-positive, −1.36 mV for ps-pH-neutral and −11.1 mV for ps-pH-negative, respectively.
3.2 Cellular internalization of the sensors
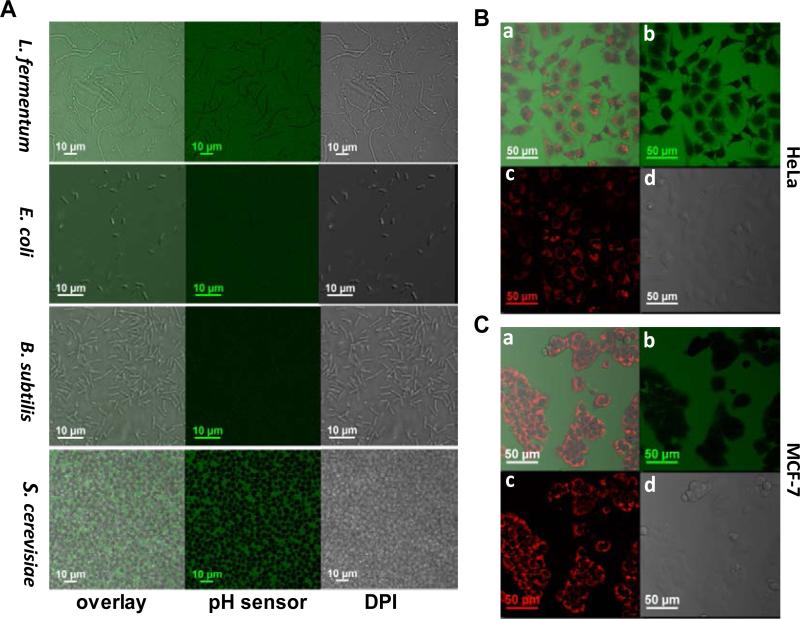
After dialysis against deionized water, the aqueous solutions of these sensors were filtered by φ0.2 μm filter before they were applied to microbial culture. Microbes were incubated with fresh medium containing 10 μg/mL of pH sensor at room temperature for at least 24h without any disturbance. No inhibition effect on cell growth was observed even after longer incubation. The distribution of sensors in microbial culture was detected by fluorescence imaging with confocal microscopy (Figure 1A and Figure S1-4). Results showed that ps-pH-neutral and ps-pH-negative were not cell permeable for any of the microbes tested in this experiment, which are Lactobacillus fermentum (gram-positive), E. coli (gram-negative), B. subtilis (gram-positive), and S. cerevisiae; while the sensor with positive charges, ps-pH-positive, is partially cell permeable for lactobacilli and yeast since weak fluorescence was observed inside cells.
Figure 1.
Distribution of pH sensor (ps-pH-neutral) when it is incubated with microbes (panel A) and mammalian cells (panels B and C). Cells were not washed in order to show the background emissions and cells were not stained by the polymer. Green emission is from the polymer excited at 488 nm, red emission (image c in panels B and C) is from MitoTracker® Red. Image a is an overlay of image b (sensor), image c (MitoTracker) and image d (DPI) in panels B and C, respectively.
A similar test was performed with mammalian cells by incubating cells with medium containing 10 μg/mL of sterilized pH sensor at 37 °C for 24h. Using MitoTracker Red as the positive control, imaging data (Figure 1B and 1C and Figure S5-7) shows that neither ps-pH-neutral or ps-pH-negative can be taken up by HeLa cell or MCF-7 cells; while the one with positive charges can get into both cell lines. It is no surprise that the macrophage cell J774 took up all three formats of polymeric pH sensors with its biological function of phagocytosis.
Based on the distribution of the above results, we selected ps-pH-neutral for further characterization and applications in measuring extracellular acidification.
3.3 Sensor (ps-pH-neutral) response to pH
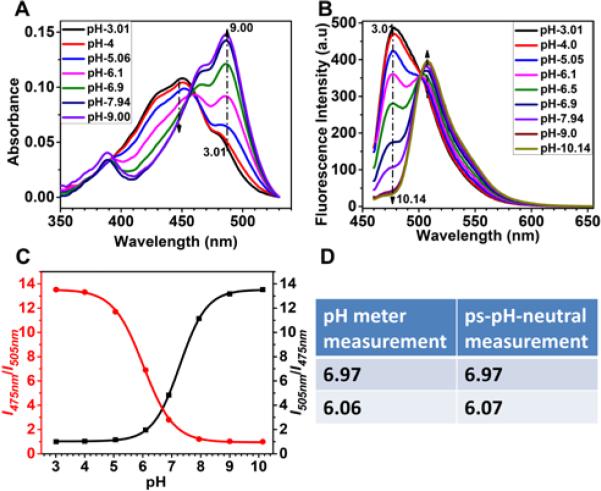
A B-R buffer with pH range 3 to 10 was applied to test the pH responses of the sensor. Similar to the sensing probe ANNA, the polymeric pH sensor shows pH-dependent absorbance spectra with an isosbestic point at 455 nm (Figure 2A) 41. The fluorescent emission spectra from the sensor under excitation at the isosbestic e point 455 nm possess two peaks at 475 nm and 505 nm, respectively (Figure 2B). The ratio of fluorescence intensity at 475 nm to the intensity at 505 nm (I475nm/I505nm or I505nm/I475nm) was plotted vs pH and the curve was fit with Bolzmann model by OriginPro 9 (Figure 2B). The curve fitting equation and the related parameters are listed in Table S1. The pH values, which were calculated from fluorescence intensity by fitting the equation of ps-pH-neutral, were validated by a pH meter. The measurement error is less than 0.01. Three concentrations of pH sensors were applied to test the concentration effects on the pH response of sensors. As shown in Figure S8, the three titration curves overlapped very well, indicating that there is not much effect from sensor concentrations on the sensor performances. 10 μg/mL of ps-pH-neutral was used for the next biological experiments.
Figure 2.
pH titration for ps-pH neutral. A: absorbance changes. B: Emission changes excited at 455 nm. C: Ratiometric plots. D: Differences for a few measurements between the results calculated from the fluorescence sensor and those from pH electrodes showing the high accuracy of measuring results using the fluorescent sensor.
3.4 Application of ps-pH-neutral in bacterial culture
The application of ps-pH-neutral was tested with three kinds of bacteria, i.e. L. fermentum, B. subtilis, and E. coli, which represent the most commonly used bacteria in their related biology and biotechnology fields (Table 1). Cuvette and 96-well microplate were applied for low throughput studies (Figure 3) and high throughput studies (Figure 4) under the sealed condition with mineral oil, respectively. In the low throughput studies, bacterial culture with pH sensor was incubated at 37 °C. The pH responding spectra in the 2 mL cuvette were collected by spectrofluorophotometer under the excitation of 455 nm (Figure 3A, 5C, and 5E). The pH responses of the sensor were also tested in a 96-well microplate to demonstrate the capacity for high throughput applications, where multiple concentrations of bacteria were repeatedly seeded in wells. The fluorescent emission at 475 nm and 505 nm from each well could be efficiently detected by a microplate reader with two-minute intervals (Figure 4A, 4C and 4E). Besides the bacteria concentration used in low throughput study (0.1 OD600), higher concentrations of bacteria (0.2, 0.3 or 0.4 OD600) were also tested in the high throughput study.
Table 1.
Bacteria used in the testing of pH sensors.
| Bacterium | Gram reaction | Metabolism | Main products of fermentation | References |
|---|---|---|---|---|
| L fermentum | positive | facultative anaerobic | lactic acid, ethanol, CO2 | 53, 54 |
| E. coli | negative | facultative anaerobic | acetate, ethanol etc. | 57-59 |
| B. subtilis | positive | facultative anaerobic | 2,3-butanediol, ethanol and acetate. | 60-63 |
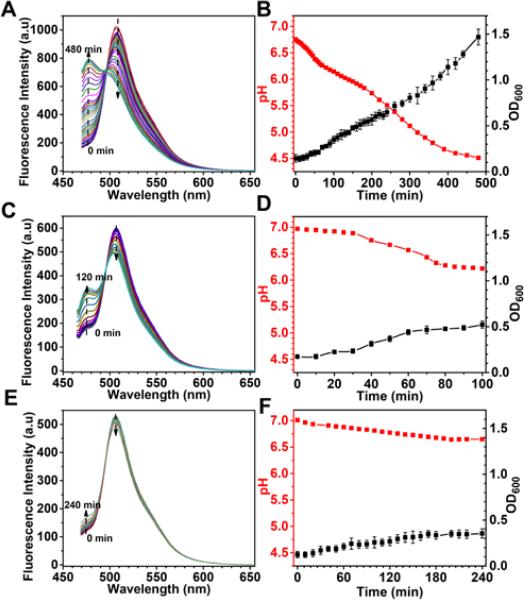
Figure 3.
Application of ps-pH-neutral in bacterial culture. Bacterial culture containing pH sensors were incubated in a sealed cuvette at 37 °C. The fluorescence spectra of pH sensors were dynamically detected by spectrofluorophotometer under 455 nm excitation, A: L. fermentum, C: E. coli, E: B. subtilis. The pH values (red) were transferred from their corresponding spectra data and were plotted together with their growth curve (black), B: L. fermentum, D: E. coli, F: B. subtilis.
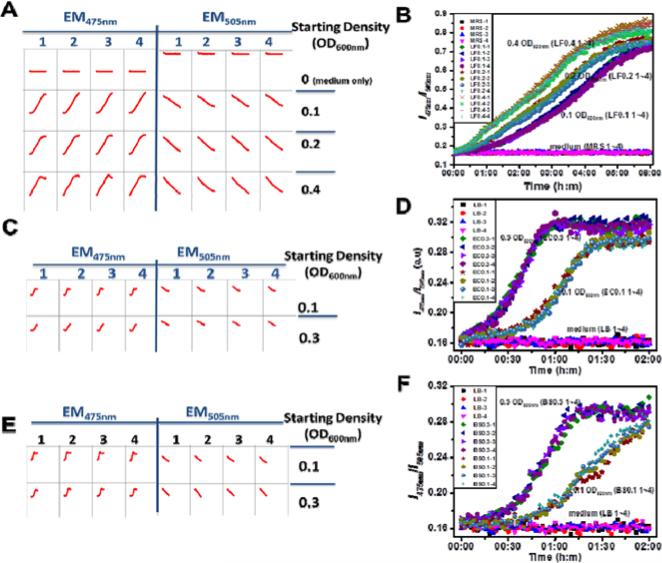
Figure 4.
Application of ps-pH-neutral in the high throughput detection of bacterial culture with microplates. Bacterial culture containing pH sensors were incubated in microplates at 37 °C. The fluorescence intensities at 475 nm and 505 nm of pH sensors were dynamically detected by a microplate reader under 455 nm excitation, A: L. fermentum, C: E. coli, E: B. subtilis. The ratio values (I475nm/I505nm) were calculated and were plotted vs. incubation time, B: L. fermentum, D: E. coli, F: B. subtilis.
Figure 5.
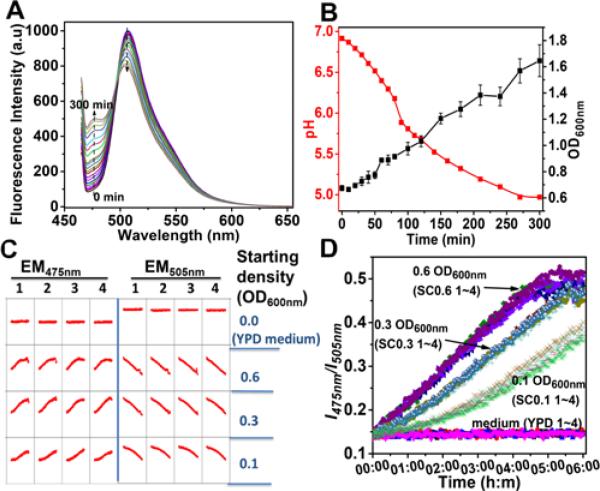
Application of ps-pH-neutral in S. cerevisiae culture. A, fluorescence spectra of pH sensor were dynamically detected by spectrofluorophotometer under 455 nm excitation. B, growth curve of yeast and pH value which were transferred from data in A were plotted vs. incubation time in a cuvette. C, screen printing data from high throughput analysis of pH in yeast culture with microplate. D, plot curves of ratio values (I475nm/I505nm) from C vs. incubation time.
L. fermentum is a Gram-positive, heterofermentative lactic acid bacterium which is wildly used in the production of fermented food 53. It was reported that the pH of L. fermentum culture dramatically affects the product of fermentation 54. It has been also used as a probiotic with its functional efficacy of antimicrobial and antioxidative activities 55, 56. During the incubation, the fluorescent intensity at 475 nm continued increasing, while the intensity at 505 nm continued decreasing (Figure 3A), meaning the cultural environment was acidized by lactobacilli under an anaerobic condition. The pH values of the cellular culture, which were calculated from the ratios of I475nm/I505nm, changed from 6.75 to 4.51 during the cell growth from 0.15 to 1.47 OD600 (Figure 3B). Three concentrations of lactobacilli, i.e. 0.1, 0.2 and 0.4 OD600, were applied in high throughput studies. As shown in Figure 4A, fluorescent emission at 475 nm and 505 nm from each well could be efficiently detected by a microplate reader. The ratio data from the same condition shows consistency throughout the experiment (Figure 4B).
E. coli is a Gram-negative, facultative anaerobe. It is the best studied bacterium and the most common bacteria used in lab cloning and also one of the preferred bacteria for research on regulation of metabolism 57, 58. Its respiratory pathways can be alternatively switched to cater to the energy request under different culture conditions. The anaerobic metabolism produces acetate, ethanol and CO2 etc. into the microenvironment. It has been reported that environmental pH affects the fermentation and plasmid product 59. The pH of E. coli culture, which was transformed from the ratio of emission peaks (Figure 3C), was changed from 6.97 to 6.22, when the density of bacteria was changed from 0.17 to 0.52 OD600 (Figure 3D). Two starting bacterial densities, i.e. 0.1 and 0.3 OD600, were used in the high throughput test (Figure 4C). During two hours of incubation at 37 °C, each well with the same condition showed a very similar ratio of fluorescence emission at 475 nm to emission at 505 nm at each time point (Figure 4D).
B. subtilis is the best studied Gram-positive facultative anaerobe that has been widely used as the paradigm of Gram-positive bacteria. The metabolism of B. subtilis, which is very sensitive to environmental pH, has been intensively investigated 60-62. B. subtilis is one of the most commonly used industrial bacterium to produce enzymes and other metabolites. The product of anaerobic fermentation of B. subtilis includes 2,3-butanediol, ethanol and acetate 63. B. subtilis were cultured under aerobic conditions. When they were seeded in an environment where the oxygen supply was blocked and residual oxygen in the medium was consumed very quickly 26, the pH of the culture did not change much, i.e. from 7.01 to 6.65 (Figure 3E and 3F). The optical density of bacteria (OD600) only grew from 0.12 to 0.35 in four hours of incubation. In the microplate, the ratios of fluorescence intensity at 455 nm and 505 nm (Figure 4E), is relatively stable and repeatable among wells with the same starting condition, 0.1 or 0.3 OD600 (Figure 4F).
Because of the different metabolites produced by bacteria under anaerobic conditions (Table 1), the pH of these three bacterial cultures changed with different trends during incubation. With the well-known capability to produce lactic acid, the pH of L. fermentum culture can reach even lower pH than 4.5 (data not shown). Under the experimental condition applied on B. subtilis, there was about 0.35 pH change detected by our pH sensor. Because of different metabolic pathways under anaerobic condition, the pH changes in different trends among three bacterial cultures tested in this experiment (Figure 4). Especially for E. coli and B. subtilis, which are normally cultured in aerobic condition, data detected by ps-pH-neutral in microplates show that the extracellular pHs changed with different rates when they were sealed in the same space with limited oxygen supply, and the same thing happened to two concentrations of the same bacteria (Figure 4D and 6F). Besides the excellent reproducibility of the sensor for high throughput applications, it is worth noting that the pH sensor shows good photo-stability during incubation, and no obvious photobleaching effect was detected (fluorescent ratio cure of medium, Figure 4B, 4D and 4F).
3.5 Application of ps-pH-neutral in S. cerevisiae culture
S. cerevisiae is the most widely used microbe in the bioindustry including the food and beverage industry, bioethanol production and other fine chemical production 64. Environmental pH has a strong effect on the growth and fermentation property of S. cerevisiae 65-67.
The application of the pH sensor in monitoring pH of S. cerevisiae culture was performed both in a cuvette and a 96-well microplate where the oxygen supply was blocked to introduce an anaerobic cell culture environment. To observe a conspicuous change of fluorescence, 0.65 OD600 starting density of yeast were sealed in the cuvette and were incubated with the pH sensor at 30 °C for five hours to detect the pH and optical density of culture, respectively (Figure 5A and 5B). As a eukaryotic organism which can grow under anaerobic conditions, the density of S. cerevisiae reached 1.65 OD600, while the pH of the culture was acidized from 6.91 to 4.97. To test the sensor response to pH in a high-throughput platform, three starting densities, 0.1, 0.3 and 0.6 OD600, of yeast were quadruplicately applied in 96-well plate (Figure 5C). During six hours of incubation, the ratios of fluorescence intensity at 475 nm and 505 nm, which were detected by microplate reader under excitation 455 nm, are well correlated with their culture conditions (Figure 5D). Signal vibration was observed at the late incubation. It might be the result of releasing CO2 from culture, which is one of the main products of fermentation with S. cerevisiae.
Besides using microbes, the ability of the pH sensor (ps-pH-neutral) to monitor extracellular pH of mammalian cells was preliminarily tested in a standard 96-well microplate. When cells grew to 90% confluence, cell culture medium in each well was substituted by medium which was adjusted to different pHs (medium-pH) containing 10 μg/mL of pH sensors (Figure S10A). The fluorescence spectra in each well were detected by a microplate reader under 440 nm, 480 nm and 455 nm excitation, respectively (Figure S10B). As we can see from fluorescence spectra in Figures S10B and S10C, each well had a good spectrum corresponding to the related pH condition. This result shows us the very promising capability of the pH sensor (ps-pH-neutral) to test multiple conditions of mammalian cells with a standard 96-well plate. No obvious cytotoxicity to mammalian cells (HeLa MCF-7 and J-774c cells, respectively) was observed after 24h of incubation with culture medium containing 10 μg/mL of the pH sensor (Figure S11).
4. Conclusion
A new water-soluble polymer-based pH sensor was developed to specifically detect extracellular pH. The varying responses of two fluorescence emission peaks (475 nm and 505 nm) under 455 nm excitation enables the sensor to ratiometrically detect pH with high accuracy. The biocompatible polymer, i.e. PHPMA, was introduced into the sensor to improve the water solubility. After 96h of incubation of cells with sensor (ps-pH-neutral), no inhibition to cell growth was observed (data not shown). Tested with Gram-negative bacteria (E. coli), Gram-positive bacteria (L. fermentum and B. subtilis), yeast (S. cerevisiae) and mammalian cells (HeLa and MCF-7), the pH sensor, ps-pH-neutral, has been proven to be exclusively extracellularly localized. This is critical to detect the environmental pH of cells. All of these characteristics ensure the pH sensor can be conveniently used to detect cell metabolism with minimal disturbance to cells.
The reliable performance in the detection of pH changes in a microplate further characterized ps-pH-neutral as a powerful tool in high-throughput screens (HTC) with a commercial microplate reader, which is widely used in drug discovery, optimization of reactions, etc. With assistance of a high resolution imaging instrument, this pH sensor has high potential to be used in single cell metabolism assays 68.
Supplementary Material
Acknowledgements
This work was supported by the NIH National Human Genome Research Institute, Centers of Excellence in Genomic Science, grant number 5 P50 HG002360, and NIH Common Fund LINCS program, grant number 5 U01 CA164250, Professor Deirdre Meldrum, PI.
Footnotes
Electronic Supplementary Information (ESI) available: See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Booth IR, Krulwich TA, Padan E, Stock JB, Cook GM, Skulachev V, Bennett GN, Epstein W, Slonczewski JL, Rowbury RJ, Matin A, Foster JW, Poole RK, Konings WN, Schafer G, Dimroth P. Novart Fdn Symp. 1999;221:19–37. [Google Scholar]
- 2.Casey JR, Grinstein S, Orlowski J. Nat Rev Mol Cell Bio. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 3.Krulwich TA, Sachs G, Padan E. Nat Rev Microbiol. 2011;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madshus IH. Biochem J. 1988;250:1–8. doi: 10.1042/bj2500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruffin VA, Salameh AI, Boron WF, Parker MD. Front Physiol. 2014;5:43. doi: 10.3389/fphys.2014.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairns RA, Harris IS, Mak TW. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 7.Gatenby RA, Gillies RJ. Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 8.Kato Y, Ozawa S, Miyamoto C, Maehata Y, Suzuki A, Maeda T, Baba Y. Cancer Cell Int. 2013;13:89. doi: 10.1186/1475-2867-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prescott LM, Harley JP, Klein DA. Microbiology. 6th edn. McGraw-Hill Higher Education; Dubuque, IA: 2005. [Google Scholar]
- 10.Thauer RK, Jungermann K, Decker K. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rofstad EK, Mathiesen B, Kindem K, Galappathi K. Cancer Res. 2006;66:6699–6707. doi: 10.1158/0008-5472.CAN-06-0983. [DOI] [PubMed] [Google Scholar]
- 12.Estrella V, Chen TA, Lloyd M, Wojtkowiak J, Cornnell HH, Ibrahim-Hashim A, Bailey K, Balagurunathan Y, Rothberg JM, Sloane BF, Johnson J, Gatenby RA, Gillies RJ. Cancer Res. 2013;73:1524–1535. doi: 10.1158/0008-5472.CAN-12-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jagielska A, Wilhite KD, Van Vliet KJ. Plos One. 2013;8:e76048. doi: 10.1371/journal.pone.0076048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L, Fukumura D, Jain RK. J Biol Chem. 2002;277:19242–19242. doi: 10.1074/jbc.M108347200. [DOI] [PubMed] [Google Scholar]
- 15.Lardner A. J Leukocyte Biol. 2001;69:522–530. [PubMed] [Google Scholar]
- 16.Teo AL, Mantalaris A, Lim M. Biochem Eng J. 2014;90:8–15. [Google Scholar]
- 17.Kim SH, Lee GM. J Microbiol Biotechn. 2007;17:712–720. [PubMed] [Google Scholar]
- 18.Arroyo-Lopez FN, Orlic S, Querol A, Barrio E. Int J Food Microbiol. 2009;131:120–127. doi: 10.1016/j.ijfoodmicro.2009.01.035. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, Zhang W, Li CJ, Sakakibara K, Tanaka S, Kong HN. Biomass Bioenerg. 2012;47:395–401. [Google Scholar]
- 20.Liu J, Wang Q, Zou H, Liu Y, Wang J, Gan K, Xiang J. Microb Biotechnol. 2013;6:685–693. doi: 10.1111/1751-7915.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piontek J, Lunau M, Handel N, Borchard C, Wurst M, Engel A. Biogeosciences. 2010;7:1615–1624. [Google Scholar]
- 22.Weber KA, Achenbach LA, Coates JD. Nat Rev Microbiol. 2006;4:752–764. doi: 10.1038/nrmicro1490. [DOI] [PubMed] [Google Scholar]
- 23.Erfle JD, Boila RJ, Teather RM, Mahadevan S, Sauer FD. J Dairy Sci. 1982;65:1457–1464. [Google Scholar]
- 24.Tian Y, Fuller E, Klug S, Lee F, Su F, Zhang L, Chao SH, Meldrum DR. Sensors and actuators. B, Chemical. 2013;188:1–10. doi: 10.1016/j.snb.2013.06.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Su F, Buizer S, Kong X, Lee F, Day K, Tian Y, Meldrum DR. Chemical communications (Cambridge, England) 2014;50:6920–6922. doi: 10.1039/c4cc01110d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Su F, Buizer S, Lu H, Gao W, Tian Y, Meldrum D. Biomaterials. 2013;34:9779–9788. doi: 10.1016/j.biomaterials.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu H, Jin Y, Tian Y, Zhang W, Holl MR, Meldrum DR. J Mater Chem. 2011;2011:19293–192301. doi: 10.1039/C1JM13754A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian Y, Su F, Weber W, Nandakumar V, Shumway BR, Jin Y, Zhou X, Holl MR, Johnson RH, Meldrum DR. Biomaterials. 2010;31:7411–7422. doi: 10.1016/j.biomaterials.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Wang Y, Yang S, Zhao Y, Yuan L, Zheng J, Yang R. Anal Chem. 2015;87:2495–2503. doi: 10.1021/ac5045498. [DOI] [PubMed] [Google Scholar]
- 30.Qi J, Liu D, Liu X, Guan S, Shi F, Chang H, He H, Yang G. Anal Chem. 2015;87:5897–5904. doi: 10.1021/acs.analchem.5b00053. [DOI] [PubMed] [Google Scholar]
- 31.Yang L, Li N, Pan W, Yu Z, Tang B. Anal Chem. 2015;87:3678–3684. doi: 10.1021/ac503975x. [DOI] [PubMed] [Google Scholar]
- 32.Nagl S, Wolfbeis OS. Analyst. 2007;132:507–511. doi: 10.1039/b702753b. [DOI] [PubMed] [Google Scholar]
- 33.Yin J, Hu Y, Yoon J. Chem Soc Rev. 2015;44:4619–4644. doi: 10.1039/c4cs00275j. [DOI] [PubMed] [Google Scholar]
- 34.Ling J, Naren G, Kelly J, Moody TS, de Silva AP. J Am Chem Soc. 2015;137:3763–3766. doi: 10.1021/jacs.5b00665. [DOI] [PubMed] [Google Scholar]
- 35.Paek K, Yang H, Lee J, Park J, Kim BJ. Acs Nano. 2014;8:2848–2856. doi: 10.1021/nn406657b. [DOI] [PubMed] [Google Scholar]
- 36.Ke G, Zhu Z, Wang W, Zou Y, Guan Z, Jia S, Zhang H, Wu X, Yang CJ. ACS Appl Mater Interfaces. 2014;6:15329–15334. doi: 10.1021/am503818n. [DOI] [PubMed] [Google Scholar]
- 37.Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A, Colone M, Tatti M, Sargiacomo M, Fais S. J Biol Chem. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peppicelli S, Bianchini F, Calorini L. Cancer Metastasis Rev. 2014;33:823–832. doi: 10.1007/s10555-014-9506-4. [DOI] [PubMed] [Google Scholar]
- 39.Valenzuela JF, Pinuer L, Cancino AG, Yanez RB. Appl Microbiol Biot. 2015;99:6417–6429. doi: 10.1007/s00253-015-6526-0. [DOI] [PubMed] [Google Scholar]
- 40.Moon C, Jang S, Yun YM, Lee MK, Kim DH, Kang WS, Kwak SS, Kim MS. Bioresource Technol. 2015;179:595–601. doi: 10.1016/j.biortech.2014.10.128. [DOI] [PubMed] [Google Scholar]
- 41.Zhou J, Fang C, Chang T, Liu X, Shangguan D. J Mater Chem B. 2013;1:661–667. doi: 10.1039/c2tb00179a. [DOI] [PubMed] [Google Scholar]
- 42.Grillo-Hill BK, Webb BA, Barber DL. Method Cell Biol. 2014;123:429–448. doi: 10.1016/B978-0-12-420138-5.00023-9. [DOI] [PubMed] [Google Scholar]
- 43.Somers RC, Lanning RM, Snee PT, Greytak AB, Jain RK, Bawendi MG, Nocera DG. Chem Sci. 2012;3:2980–2985. doi: 10.1039/C2SC20212C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin T, Sasaki A, Kinjo M, Miyazaki J. Chem Commun. 2010;46:2408–2410. doi: 10.1039/b921602b. [DOI] [PubMed] [Google Scholar]
- 45.Long L, Li X, Zhang D, Meng S, Zhang J, Sun X, Zhang C, Zhou L, Wang L. Rsc Adv. 2013;3:12204–12209. [Google Scholar]
- 46.Cannon TM, Shah AT, Walsh AJ, Skala MC. J Biomed Opt. 2015:20. doi: 10.1117/1.JBO.20.1.010503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.An WF, Tolliday N. Mol Biotechnol. 2010;45:180–186. doi: 10.1007/s12033-010-9251-z. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Y, Hu Q, Cheng F, Su N, Wang A, Zou Y, Hu H, Chen X, Zhou HM, Huang X, Yang K, Zhu Q, Wang X, Yi J, Zhu L, Qian X, Chen L, Tang Y, Loscalzo J, Yang Y. Cell Metab. 2015;21:777–789. doi: 10.1016/j.cmet.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Musken M, Di Fiore S, Romling U, Haussler S. Nat Protoc. 2010;5:1460–1469. doi: 10.1038/nprot.2010.110. [DOI] [PubMed] [Google Scholar]
- 50.Lu H, Su F, Mei Q, Tian Y, Tian W, Johnson RH, Meldrum DR. J Mater Chem. 2012;22:9890–9900. doi: 10.1039/C2JM30258F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelsch A, Tomcin S, Rausch K, Barz M, Mailander V, Schmidt M, Landfester K, Zentel R. Biomacromolecules. 2012;13:4179–4187. doi: 10.1021/bm301453g. [DOI] [PubMed] [Google Scholar]
- 52.Lammers T, Ulbrich K. Adv Drug Deliver Rev. 2010;62:119–121. doi: 10.1016/j.addr.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 53.Jayashree S, Pooja S, Pushpanathan M, Vishnu U, Sankarasubramanian J, Rajendhran J, Gunasekaran P. Genome Announc. 2013;1:e00770–13. doi: 10.1128/genomeA.00770-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santoyo MC, Loiseau G, Sanoja RR, Guyot JP. Int J Food Microbiol. 2003;80:77–87. doi: 10.1016/s0168-1605(02)00140-x. [DOI] [PubMed] [Google Scholar]
- 55.Mikelsaar M, Zilmer M. Microb Ecol Health Dis. 2009;21:1–27. doi: 10.1080/08910600902815561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reid G. Appl Environ Microbiol. 1999;65:3763–3766. doi: 10.1128/aem.65.9.3763-3766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dharmadi Y, Murarka A, Gonzalez R. Biotechnol Bioeng. 2006;94:821–829. doi: 10.1002/bit.21025. [DOI] [PubMed] [Google Scholar]
- 58.Unden G, Bongaerts J. Biochim Biophys Acta. 1997;1320:217–234. doi: 10.1016/s0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 59.Cortes JT, Flores N, Bolivar F, Lara AR, Ramirez OT. Biotechnol Bioeng. 2015;113:598–611. doi: 10.1002/bit.25817. [DOI] [PubMed] [Google Scholar]
- 60.Sonenshein AL. Nat Rev Microbiol. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- 61.Buescher JM, Liebermeister W, Jules M, Uhr M, Muntel J, Botella E, Hessling B, Kleijn RJ, Le Chat L, Lecointe F, Mader U, Nicolas P, Piersma S, Rugheimer F, Becher D, Bessieres P, Bidnenko E, Denham EL, Dervyn E, Devine KM, Doherty G, Drulhe S, Felicori L, Fogg MJ, Goelzer A, Hansen A, Harwood CR, Hecker M, Hubner S, Hultschig C, Jarmer H, Klipp E, Leduc A, Lewis P, Molina F, Noirot P, Peres S, Pigeonneau N, Pohl S, Rasmussen S, Rinn B, Schaffer M, Schnidder J, Schwikowski B, Van Dijl JM, Veiga P, Walsh S, Wilkinson AJ, Stelling J, Aymerich S, Sauer U. Science. 2012;335:1099–1103. doi: 10.1126/science.1206871. [DOI] [PubMed] [Google Scholar]
- 62.Wilks JC, Kitko RD, Cleeton SH, Lee GE, Ugwu CS, Jones BD, BonDurant SS, Slonczewski JL. Appl Environ Microb. 2009;75:981–990. doi: 10.1128/AEM.01652-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cruz Ramos H, Hoffmann T, Marino M, Nedjari H, Presecan-Siedel E, Dreesen O, Glaser P, Jahn D. J Bacteriol. 2000;182:3072–3080. doi: 10.1128/jb.182.11.3072-3080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nevoigt E. Microbiol Mol Biol R. 2008;72:379–412. doi: 10.1128/MMBR.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu X, Jia B, Sun X, Ai J, Wang L, Wang C, Zhao F, Zhan J, Huang W. J Food Sci. 2015;80:M800–808. doi: 10.1111/1750-3841.12813. [DOI] [PubMed] [Google Scholar]
- 66.Nielsen MK, Arneborg N. Food Microbiol. 2007;24:101–105. doi: 10.1016/j.fm.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 67.Pena A, Sanchez NS, Alvarez H, Calahorra M, Ramirez J. Fems Yeast Res. 2015;15:fou005. doi: 10.1093/femsyr/fou005. [DOI] [PubMed] [Google Scholar]
- 68.Lidstrom ME, Meldrum DR. Nat Rev Microbiol. 2003;1:158–164. doi: 10.1038/nrmicro755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.