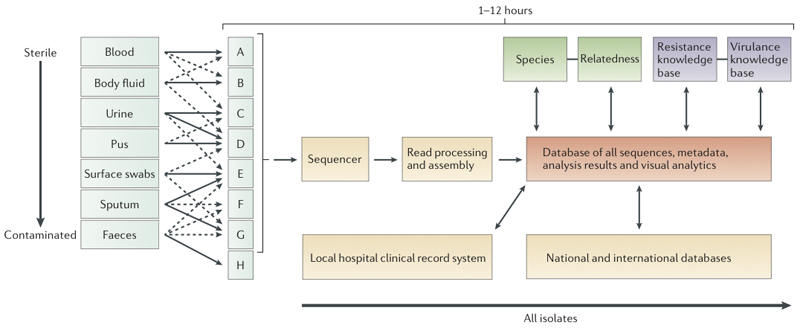
Figure 2. Hypothetical workflow based on whole genome sequencing.
Schematic representation of the workflow anticipated after adoption of whole genome sequencing, with an expected timescale that could fit within a single day. The culture steps would be the same as currently used in a routine microbiology laboratory. Some types of sample might be directly sequenced (see ‘future directions’, not shown here). Once a sample or likely pathogen is ready for sequencing, DNA will be extracted. This procedure is becoming simpler, as the input required for successful sequencing is reducing; it is now possible to use as little as 5 ng and to purify this in <30 minutes. For current bench-top machines it can take as little as 2 hours to prepare the DNA for sequencing, and new platforms (Box 1) could enable sequencing without preparation. Therefore, bacterial genome sequencing in hours and possibly even minutes is a realistic prospect.
After sequencing, the main processes for yielding information will be computational. The development of software and databases is a major challenge to overcome before a pathogen sequencing can be deployed in clinical microbiology. Automated sequence assembly algorithms will be necessary for processing the raw sequence data (Box 1). This assembled sequence would then be analysed by modular software to determine species, relationship to other isolates of the same species, antimicrobial resistance profile and virulence gene content. Results of this analysis will be reported through hospital information systems. All the results will also be used for outbreak detection and infectious diseases surveillance. These developments will require new large database and other informatics technology and will take time to develop. In particular, it will need ‘intelligent systems’ which will incorporate elements of machine learning to enable automatic updating of key knowledge bases for species identification, antimicrobial resistance determination and virulence detection. Formal evaluation of such a solution will also need robust testing to ensure it performs at least as well as current methods.