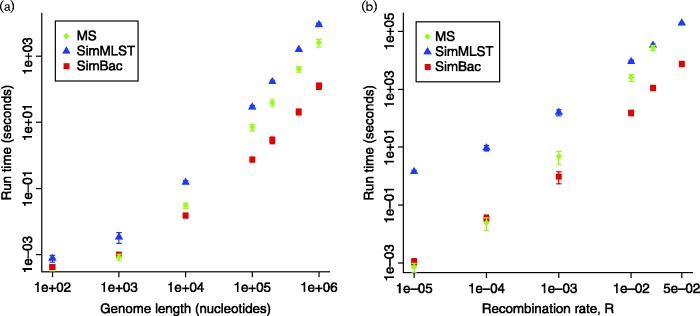
Fig. 1.
Comparison of run-time of SimMLST, ms and SimBac. Only gene conversion (no crossover) is simulated in ms, to model bacterial evolution. (a) Mean time to simulate the ARG for a fixed recombination rate R = 0.01 and genome length from 100 bp to 1 Mbp. (b) Mean time to simulate the ARG for a fixed genome length of 1 Mbp and recombination rate increasing from R = 0 to R = 0.05. One hundred simulations were performed for each dot, except for SimMLST at R = 0.02 and R = 0.05, and ms at R = 0.02, where 10 simulations were performed due to the elevated computational demand. ms was not run at R = 0.05 because a single run required >4 days. Error bars show ± 1 sd.