SUMMARY
Perception of microbe-associated molecular patterns (MAMPs) by cell surface-resident pattern recognition receptors (PRRs) induces rapid, robust and selective transcriptional reprogramming, which is central for launching effective pattern-triggered immunity (PTI) in plants. Signal relay from PRR complexes to the nuclear transcriptional machinery via intracellular kinase cascades rapidly activates primary immune response genes. The coordinated action of gene-specific transcription factors and the general transcriptional machinery contributes to the selectivity of immune gene activation. In addition, PRR complexes and signaling components are often transcriptionally up-regulated upon MAMP perception to ensure the robustness and sustainability of PTI outputs. In this review, we discuss recent advances in deciphering the signaling pathways and regulatory mechanisms that coordinately lead to timely and accurate MAMP-induced gene expression in plants.
Keywords: Microbe-associated molecular patterns (MAMPs), pattern-triggered immunity (PTI), Transcriptional reprogramming, transcription factors, general transcriptional machinery
INTRODUCTION
Being sessile and photoautotrophic, plants need to launch prompt and effective defense responses in situ to ward off constant threats from diverse pathogens. Plants also need to precisely coordinate defense with growth and development to optimize ecological fitness. Plants defend against pathogens using a two-tiered innate immune system: pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) (Jones and Dangl, 2006). PTI is triggered via activation of plasma membrane (PM)-resident pattern recognition receptors (PRRs) by pathogen- or microbe-associated molecular patterns (PAMPs/MAMPs) and endogenous damage-associated molecular patterns (DAMPs) (Macho and Zipfel, 2014). ETI is elicited by pathogen-derived effectors via intracellular nucleotide-binding domain leucine-rich repeat-containing receptors (NLRs) and is often accompanied with localized programmed cell death (Zebell and Dong, 2015). In general, PTI contributes to plant basal resistance to diverse adapted and non-adapted microbes, whereas ETI plays a central role in defending against race-specific host-adapted pathogens. In addition, localized primary infection can signal the development of systemic acquired resistance, which manifests as enhanced resistance in distal tissues (Spoel and Dong, 2012).
Pathogen invasion usually induces a profound and dynamic reprogramming of plant gene expression, which is central for launching robust and effective host defense responses (Buscaill and Rivas, 2014; Tsuda and Somssich, 2015). Recent studies have substantially advanced our understanding of the genome-wide transcriptional landscape underlying plant defense responses and uncovered roles for some pathway-specific transcription factors in regulating plant immune gene expression. Further, along with the uncovering of pathways from pathogen perception to gene activation, the specific functions of general transcriptional machinery in regulating primary immune gene expression are emerging. Here, we review the recent progress on the mechanisms of MAMP-induced transcriptional reprogramming and transcriptional regulation of PTI outputs in plants. We aim to provide insights into how the cooperative action of the general transcriptional machinery and specific transcription factors is orchestrated to achieve rapid and highly selective activation of immune genes, and how transcriptional regulation sustains various PTI outputs.
Robust and dynamic transcriptional changes in PTI
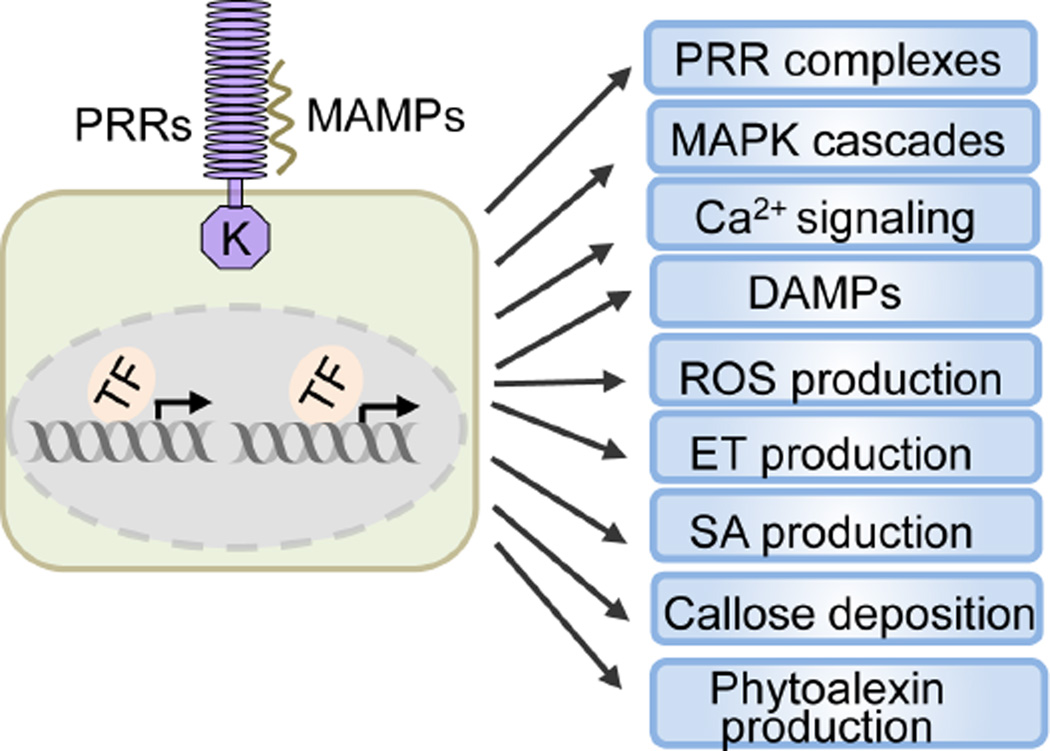
The well-characterized Arabidopsis PRRs include the bacterial flagellin receptor FLS2 that recognizes a conserved 22-amino-acid peptide (flg22) of flagellin and the bacterial elongation factor Tu (EF-Tu) receptor EFR that recognizes an 18-amino-acid peptide (elf18) of EF-Tu. Both FLS2 and EFR are leucine-rich repeat receptor-like kinases (LRR-RLKs). Upon perception of the cognate MAMPs, these PRRs initiate immune signaling by instantaneous heterodimerization with the LRR-RLK family co-receptor BAK1 (Bohm et al., 2014; Macho and Zipfel, 2014). The receptor-like cytoplasmic kinase (RLCK) BIK1 and its homolog PBL1 constitutively associate with FLS2/EFR and BAK1, and are rapidly phosphorylated and released from the receptor complexes upon MAMP perception (Lin et al., 2013). BIK1 directly phosphorylates the PM-resident NADPH oxidase RBOHD, which together with calcium signaling-mediated RBOHD regulation results in the transient production of reactive oxygen species (ROS) (Macho and Zipfel, 2014). Perception of MAMPs also activates mitogen-activated protein kinases (MAPKs) and calcium-dependent protein kinases (CDPKs), two central signaling modules that transduce early PTI signals into multiple intracellular defense responses including transcriptional reprogramming (Tena et al., 2011). Callose deposition, stomatal closure, and production of ethylene (ET) and antimicrobial components are the resulting hallmarks of PTI, which are regulated downstream or independently of MAPK and CDPK activations (Figure 1) (Macho and Zipfel, 2014; Wu et al., 2014). Collectively, these events lead to the inhibition of pathogen multiplication and containment of disease progression. Further, genes encoding DAMPs, PRR complexes and signaling components are often rapidly and transiently upregulated in response to MAMP perception leading to the amplification of innate immune responses.
Figure 1. Plant PTI signaling and outputs are regulated by transcription.
Perception of different MAMPs by the cognate PRRs controls various PTI responses via transcriptional regulation. TF: transcription factor.
Perception of different MAMPs by cognate PRRs triggers profound transcriptional reprogramming of a set of largely overlapping genes with distinct temporal dynamics. A large number of genes are commonly upregulated by treatments with the MAMPs flg22, elf18, bacterial peptidoglycan or fungal chitin (Gust et al., 2007; Wan et al., 2008; Zipfel et al., 2006). In addition, treatment with the DAMP oligogalacturonides (OGs), pectin fragments released from plant cell walls by fungal polygalacturonases, activates a similar early transcriptional reprogramming as that induced by flg22 (Denoux et al., 2008). Thus, diverse MAMPs/DAMPs likely activate the convergent signaling pathways that lead to similar transcriptional changes. However, the induction dynamics and amplitude of individual genes may differ in response to different MAMPs.
The transcriptional reprogramming activated by the bacterium Pseudomonas syringae pv. tomato (Pst) type III secretion mutants, such as hrpA or hrcC, is likely due to a collective action of multiple MAMPs in these nonpathogenic bacteria. Comparative analysis of Arabidopsis transcriptional changes in response to virulent Pst and its nonpathogenic hrpA mutant at different time points has revealed that the transcriptional response to MAMPs is initiated prior to pathogen multiplication. The genes induced early are related to defense responses and salicylic acid (SA) biosynthesis, whereas genes associated with photosynthesis-related processes are significantly suppressed, suggesting that plants may actively reduce the production of photosynthates to restrict the resources required for pathogen growth as an additional defense mechanism (Lewis et al., 2015).
MAPKs and CDPKs are potential convergent points downstream of PRRs in controlling PTI transcriptional reprogramming (Tena et al., 2011). MAPKs and CDPKs act either specifically or synergistically in controlling MAMP-induced genes (Boudsocq et al., 2010). Perception of MAMPs rapidly and transiently activates three major MAPKs, MPK3, MPK4 and MPK6, which have redundant and distinct functions in PTI signaling. A comparative transcriptome analysis of the mpk3, mpk4, and mpk6 mutants revealed that 36% of the flg22-upregulated genes and 68% of the flg22-downregulated genes were affected in at least one of the mpk mutants (Frei dit Frey et al., 2014). A large portion of MAPK target genes are similarly regulated by two or three MAPKs, consistent with their partially overlapping functions. Identification of specific transcription factors targeted by individual MAPKs and CDPKs will provide insights into the interplay between MAPKs and CDPKs on immune gene regulation.
The plant defense hormones SA and ET/jasmonic acid (JA) are thought to function antagonistically in plants against biotrophic and necrotrophic pathogens. Analysis of an Arabidopsis dde2/ein2/pad4/sid2 quadruple mutant, in which four signaling sectors including JA, ET, phytoalexin-deficient 4 (PAD4) and SA signaling pathways are all disrupted, revealed that all three hormones and the PAD4 sector act positively in flg22/elf18-mediated PTI (Tsuda et al., 2009). Although 80% of MAMP-mediated resistance is abolished in the quadruple mutant, the early MAMP signaling events, such as MAPK activation and primary immune gene induction, were not significantly changed in the quadruple mutant, suggesting that the signaling network defined by SA, JA, ET and PAD4 sectors mainly controls some of the late PTI responses (Tsuda et al., 2009). In a model-based analysis using different combinatorial mutants of these signaling sectors, it has been shown that the interaction between SA and PAD4 sectors exerts synergistic contributions to PTI, whereas the other interactions were compensatory (Kim et al., 2014). The inhibitory effect of the ET sector on the JA sector plays an important role to the network robustness, ensuring that the network output does not change much when conditions of the network change. This network robustness is likely important for plant defense against attacks by fast-evolving pathogens (Kim et al., 2014). In addition, JA positively regulates SA, and the negative interaction between JA and PAD4 is likely the cause of apparent SA-JA antagonism. Future systems and modeling analysis will likely provide novel insights to explain the sophisticated and complicated plant immune system.
Selective activation of the general transcription machinery in plant PTI
Transcription of eukaryotic messenger RNA (mRNA) is catalyzed by the multi-subunit enzyme RNA polymerase II (RNAPII) that binds selectively to the promoters of target genes with the assistance of general transcription factors. The Mediator complex functions as transcription cofactors to regulate RNAPII activities at each step of transcription cycles and contributes to transcriptional specificity via interaction with gene-specific transcription factors that cooperatively activate target genes by binding to the specific cis-elements in promoters (Figure 2). The initiation and elongation of mRNA transcripts are also regulated by a series of additional co-activators or co-repressors, in particular those regulating histone modification and chromatin remodeling. The transcriptional responses to pathogen infections are fine-tuned at multiple levels via highly sophisticated regulatory mechanisms.
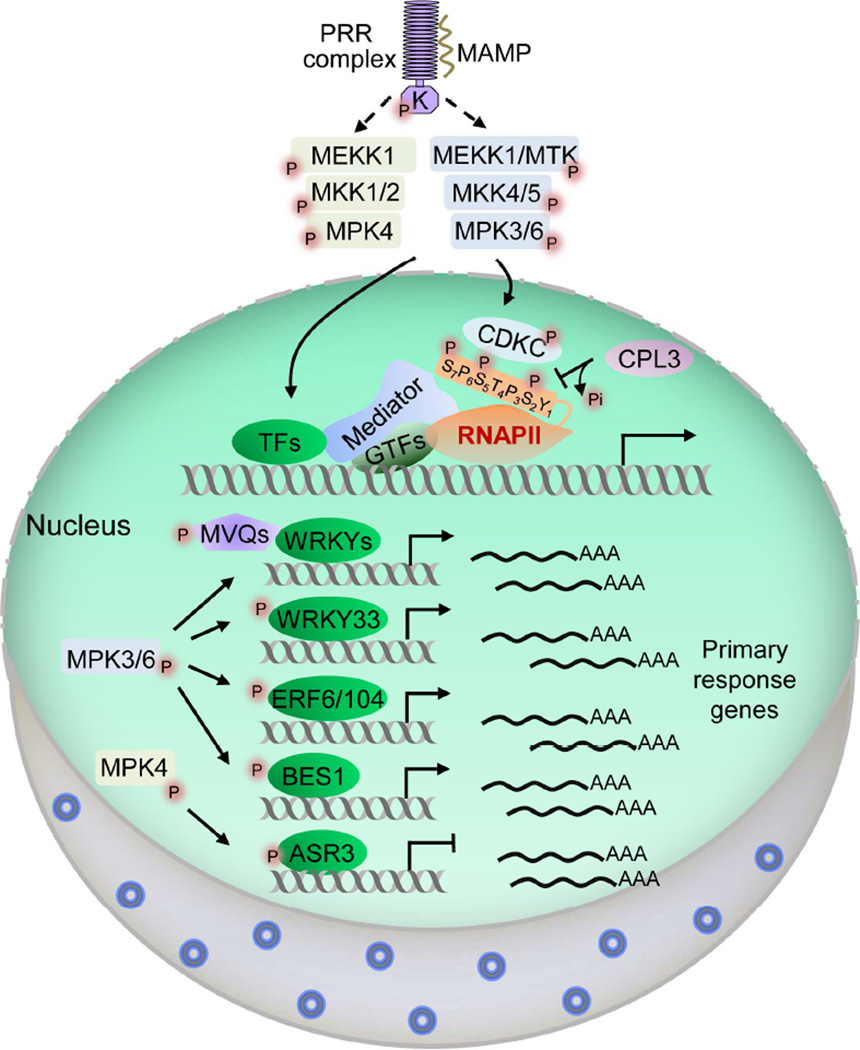
Figure 2. Phosphorylation relays control nuclear transcription dynamics in plant PTI.
Transcription of eukaryotic mRNA is controlled by the concerted action of RNAPII, general transcription factors (GTFs), Mediator, gene-specific transcription factors (TFs) and other transcription regulators, many of which are regulated by MAMP-activated MAPK cascades. Perception of MAMPs activates two MAPK cascades: MEKK1/MTK-MKK4/MKK5-MPK3/MPK6 and MEKK1-MKK1/MKK2-MPK4. MPK3/MPK6 directly phosphorylate CDKCs, which in turn phosphorylate CTD of RNAPII. CTD phosphorylation is counter-regulated by the CPL3 phosphatase. MAPKs also phosphorylate individual TFs to regulate their activities or stability. For instance, MPK3/MPK6 phosphorylate WRKY33, ERF6/ERF104 and BES1, whereas MPK4 phosphorylates ASR3 to control the expression of a subset of MAMP responsive genes. In some cases, MPK3/MPK6 directly phosphorylate MVQs, which interact with different WRKYs. Individual Mediator subunits interact with different pathway-specific TFs and contribute to the selectivity of immune gene activation. P indicates phosphorylation modification.
Phosphorylation dynamics of RNAPII in plant PTI signaling
Rpb1, the largest subunit of RNAPII, contains a carboxyl-terminal domain (CTD) consisting of conserved heptapeptide repeats with the consensus sequence Y1S2P3T4S5P6S7. Dynamic phosphorylation pattern of the CTD regulates each step of RNAPII-mediated transcription cycles via recruiting different CTD binding proteins, and thus plays a central role in the regulation of gene expression (Buratowski, 2009). As a core regulatory mechanism of eukaryotic transcription, Arabidopsis RNAPII CTD phosphorylation is deliberately regulated upon MAMP perception to launch prompt host immune responses (Li et al., 2014b). Perception of flg22, elf18 or chitin leads to a rapid and transient phosphorylation of Arabidopsis RNAPII at various serine residues in the CTD heptapeptide repeats by Arabidopsis cyclin-dependent kinase Cs (CDKCs), homologs of mammalian CDK9. MAMP-stimulated MPK3/MPK6 directly phosphorylate and activate CDKCs, which in turn phosphorylate the CTD of RNAPII (Figure 2). Consistent with this, flg22-induced CTD phosphorylation is compromised in cdkc mutants, accompanied by reduced immune gene induction and disease resistance, suggesting that CTD phosphorylation positively regulates plant PTI (Li et al., 2014b). CDKCs are also important regulators in plant virus resistance likely through regulation of viral replication in host cells (Cui et al., 2007). CDKCs appear to have a general role in gene transcription since they phosphorylate all three serine residues in CTD and are also essential for plant growth (Cui et al., 2007; Li et al., 2014b). Direct phosphoregulation of CDKCs by MAMP-activated MAPKs provides a means for plants to rapidly activate primary immune response genes.
CTD phosphorylation is intricately intertwined with dephosphorylation by different CTD phosphatases during transcription cycles. Arabidopsis CTD phosphatase-like 3 (CPL3) preferentially interacts with and dephosphorylates serine 2 of phospho-CTD (Li et al., 2014b). CPL3 negatively regulates plant PTI through counter-regulation of MAMP-induced CTD phosphorylation. Interestingly, although CPL3 regulates a significant proportion of flg22-induced genes, it barely affects the overall transcriptional profile in the absence of a stimulus. In addition, CPL3 is dispensable for Arabidopsis vegetative growth, which is distinct from the essential function of its yeast homolog FCP1 in growth regulation. This apparent paradox could be due in part to the existence of another Arabidopsis FCP1 homolog CPL4, an essential gene for plant growth and development (Bang et al., 2006). Furthermore, CPL3 is involved in plant responses to abiotic stresses mediated by the hormone abscisic acid (ABA) (Jiang et al., 2013). It is plausible that CPL4 regulates RNAPII activity for general transcription, whereas CPL3-mediated RNAPII dephosphorylation is specifically engaged in plant stress and immune responses. Notably, unlike CPL4, CPL3 has a plant-specific N-terminal domain of unknown function, which may interact with signal-specific transcriptional regulatory proteins, thereby orchestrating functional selectivity.
Differential functions of the Mediator complex in plan immunity
As an RNAPII cofactor, Mediator is an evolutionarily conserved multiunit protein complex interacting with RNAPII, other cofactors and gene-specific transcription factors in eukaryotes (Kidd et al., 2011) (Figure 2). The Mediator complex not only functions in modulation of basal transcriptional activity but also controls selective activation of specific genes in responses to various stimuli. The Arabidopsis Mediator complex consists of 21 conserved and 6 plant-specific subunits. It appears that individual Mediator subunits are engaged in diverse biological processes, suggesting their potential involvement in transducing specific signals from distinct internal and external cues to RNAPII for selective transcription of pathway-specific genes (Kidd et al., 2011). Several Arabidopsis Mediator subunits have been implicated in plant immunity to different biotrophic and necrotrophic pathogens through regulation of gene transcription. MED8, MED16, MED21 and MED25 are involved in JA/ET-mediated resistance to the necrotrophic fungi Botrytis and Alternaria (Kidd et al., 2011; Samanta and Thakur, 2015). MED18 is also important for plant immunity against necrotrophic fungi, but in a JA/ET-independent manner (Lai et al., 2014). MED14, MED15 and MED16 contribute to SA-mediated plant local and systemic resistance to Pst (Zhang et al., 2013). CDK8, MED12 and MED13, the kinase module of the Mediator complex, also play important roles in plant immunity to necrotrophic fungi and Pst (Zhu et al., 2014). Systematic characterization of various PTI outputs triggered by purified MAMPs or Pst hrcC in individual Mediator complex mutants can help reveal whether and how Mediator directly conveys PRR signaling to selective activation of MAMP-responsive genes. Consistent with its important role in regulating immune gene expression, Mediator is targeted by pathogen effectors to dampen host immunity. The secreted effector HaRxL44 of downy mildew Hyaloperonospora arabidopsidis interacts with and degrades the Arabidopsis MED19a, a positive regulator in SA-mediated immunity, to promote pathogenicity (Caillaud et al., 2013).
Individual Mediator subunits often interact with gene-specific transcription factors to decode information from upstream signals and dictate pathway-specific gene transcription. MED25 directly interacts with several key transcription factors in the JA signaling pathway, including ERF1, ORA59 and MYC2, and positively regulates JA-responsive genes (Cevik et al., 2012; Chen et al., 2012). Notably, ERF1 and ORA59 regulate different JA-responsive genes from those regulated by MYC2 (Cevik et al., 2012). In addition, MED25 as well as its interacting subunit CDK8 also regulate the expression of ERF1 and ORA59 by binding to their promoters (Cevik et al., 2012; Zhu et al., 2014). Moreover, MED16 directly interacts with WRKY33, a member of the WRKY family of plant-specific transcription factors (discussed below in detail) and is essential for the recruitment of RNAPII to the WRKY33 target genes PDF1.2 and ORA59, suggesting a specific role of MED16 in mediating the transcriptional regulation of WRKY33 target genes (Wang et al., 2015). MED18 also interacts with multiple transcription factors involved in defense responses, flowering time control and ABA signaling. MED18 is important for the RNAPII occupancy of the promoter, coding and terminator regions of target genes, reinforcing the role of Mediator in facilitating RNAPII-mediated transcription initiation, elongation and termination (Lai et al., 2014). The understanding of how a specific signaling pathway regulates the interaction of gene-specific transcription factors with Mediator subunits and the subsequent activation of transcriptional machinery may lead to the strategic development for selective activation of defense responses.
Other transcriptional co-activators in plant PTI
In addition to Mediator, many other transcriptional co-activators promote transcription by bridging transcription factors and RNAPII and/or relaxing the chromatin structure at target gene promoters (Ding and Wang, 2015). Several subunits of general transcription factors, such as TAFII-250, a major subunit of the transcription initiation factor TFIID, possess histone acetyltransferase activity. Histone acetylation of a target chromatin region is generally correlated with active transcription. The CREB-binding protein (CBP)/p300 family, a group of histone acetyltransferases in animals, are key transcriptional co-activators in gene transcriptional regulation by promoting acetylation of both histones and transcription factors. The Arabidopsis histone acetyltransferase HAC1, a homolog of CBP/p300, is involved in priming of PTI responses triggered by hrcC or flg22 (Singh et al., 2014). HAC1 is required for repetitive abiotic stress-induced plant resistance to Pst infections and enhanced PTI marker gene expression, which is correlated with RNAPII enrichment and histone modification marks associated with transcriptional activation. HAC1 is also important for priming of flg22 or hrcC-induced callose deposition in response to repetitive stresses. This may be achieved through regulation of callose biosynthetic genes, many of which are MAMP inducible (Clay et al., 2009). Apparently, the HAC1-mediated histone modification is transient and the enhanced resistance induced by repetitive stresses was lost seven days after the last stress treatment. Despite pleiotropic growth defects, without repetitive stress treatments, the hac1 mutants display no detectable difference from wild-type plants in bacterial resistance, suggesting that HAC1 is less important for plant immunity under normal conditions (Singh et al., 2014).
The ATP-dependent chromatin remodeling complex SWR1 is also involved in plant immunity and PTI signaling. SWR1 complex regulates chromatin structure by substitution of canonical H2A histone with the histone variant H2A.Z (March-Diaz et al., 2008). The Arabidopsis SWR1 complex components SEF, ARP6 and PIE1, the homologues of the yeast Swc6, Arp6 and the core ATPase Swr1 proteins respectively, negatively regulate resistance to virulent Pst and SA-mediated defense (Cheng et al., 2013; March-Diaz et al., 2008). Similarly, mutation of H2A.Z coding genes HTA9 and HTA11 also renders plants more resistant to Pst infections (March-Diaz et al., 2008). Interestingly, Arabidopsis arp6 and hta9hta11 mutants display differential responses in PTI and ETI with enhanced flg22-induced MAPK activation and PTI marker gene expression, but reduced bacterial effector-triggered hypersensitive response and resistance (Cheng et al., 2013). It has been reported that histone H2A.Z nucleosomes may function as a thermosensor for Arabidopsis to precisely perceive ambient temperature (Kumar and Wigge, 2010). The arp6 and hta9hta11 mutants phenocopy plants grown at the elevated ambient temperature. The differential responses of arp6 and hta9hta11 mutants in two tiers of plant immunity is consistent with the observation that elevated ambient temperatures promote PTI but suppress ETI (Cheng et al., 2013). Nevertheless, SWR1 complex and H2A.Z nucleosomes sense temperature and regulate plant immunity likely through modulation of transcriptome changes. In addition, other histone modification enzymes and chromatin remodeling complex components have also been implicated in plant defense to different biotrophic and necrotrophic pathogens (Ding and Wang, 2015; Ma et al., 2011). However, the knowledge on whether and how these components regulate MAMP-induced gene expression and other PTI responses awaits further investigation.
Activation of gene-specific transcription factors in PTI signaling
Transcriptional selectivity is achieved by coordinated actions of gene-specific transcription factors that bind unique DNA sequences and selectively regulate the expression of gene subsets in response to specific stimuli. Arabidopsis and other plant genomes encode ~2000 putative transcription factor genes grouped into more than 70 families. Some plant-specific families of transcription factors play critical roles in orchestrating plant immune gene expression. In general, these transcription factors often act downstream of MAPK cascades or Ca2+ signaling via diverse activation mechanisms in response to pathogen infections (Figures 2 & 3).
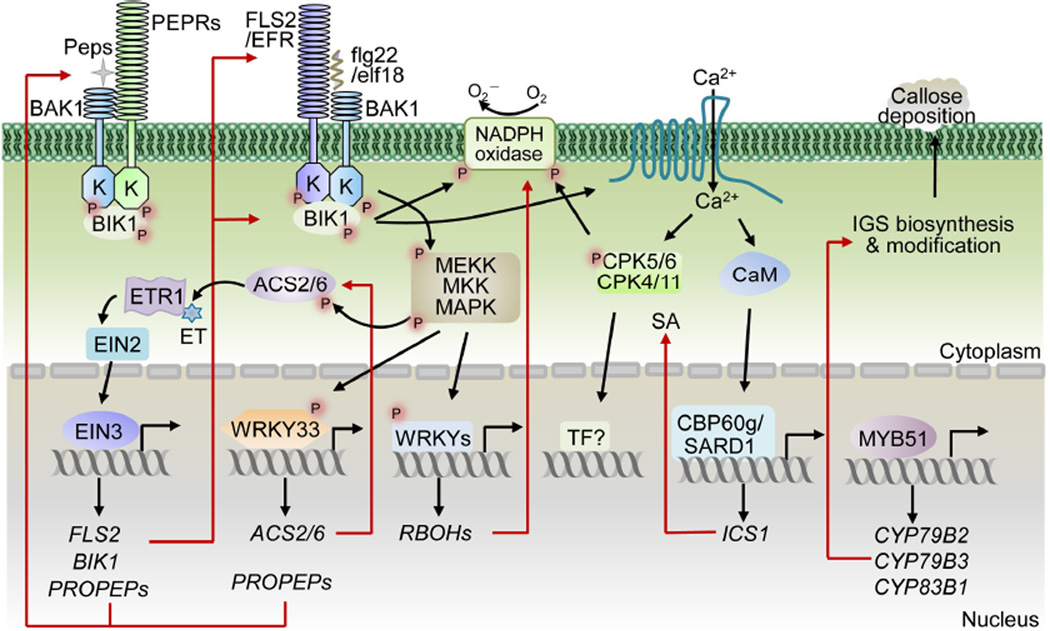
Figure 3. Transcriptional control of plant PTI signaling components and outputs.
MAMPs, flg22 and elf18, are perceived by the cognate PRRs, FLS2 and EFR, and induce PRR heterodimerization with co-receptor BAK1 family RLKs to phosphorylate and activate BIK1 family RLCKs. MAPKs and Ca2+ signaling are two convergent points downstream of multiple PRRs. MAPKs can phosphorylate some WRKYs, which control the expression of RBOHs encoding NADPH oxidase for ROS production. MPK3/MPK6 control ET production through stabilization of ACS2/ACS6 by phosphorylation and transcriptional activation of ACS2/ACS6 via phosphorylated WRKY33. WRKY33 also regulates expression of PROPEPs to amplify the danger signals via Pep-PEPR recognition. Perception of ET by ETR1 activates the TF EIN3 via EIN2. EIN3 controls the expression of FLS2 and BIK1. ET is also important for the MAMP-induced expression of PROPEPs. MYB51 encodes a TF important for callose deposition via regulating genes for precursor IGS biosynthesis. Downstream of Ca2+ signaling, several CaM-binding protein CBP60 family members regulate MAMP-induced SA accumulation via binding to the SA biosynthetic gene ICS1 promoter. Red lines and arrows indicate the transcriptional regulation of PTI responses.
Transcription factors regulated by MAPK cascades
Members of the WRKY gene family of plant-specific transcription factors are often induced in response to diverse pathogens and abiotic stresses, suggesting their broad involvement in stress responses (Tsuda and Somssich, 2015). WRKY22 and WRKY29 are induced by flg22 treatment and function downstream of a flg22-activated MAPK cascade consisting of MEKK1/MTK-MKK4/MKK5-MPK3/MPK6 (Asai et al., 2002), although the mechanism by which WRKY22/WRKY29 are activated downstream of MPK3/MPK6 is not known. On the other hand, MPK3/MPK6 directly phosphorylates WRKY33 to modulate the expression of key genes for the biosynthesis of camalexin, a major phytoalexin in Arabidopsis defense against the necrotrophic fungus Botrytis (Mao et al., 2011). WRKY33 also directly targets ABA biosynthetic genes to repress the biosynthesis of ABA, which plays a negative role in plant resistance to Botrytis (Liu et al., 2015). WRKYs often form positive feedback regulatory loops via binding to their own promoters to amplify the signal (Liu et al., 2015; Mao et al., 2011). In other cases, VQ motif-containing proteins (VQPs) interact with and bridge MAPKs and WRKYs. Flg22-activated MPK4 phosphorylates the VQP MKS1, which leads to the subsequent dissociation of the MKS1-WRKY33 complex from MPK4 to regulate target gene expression (Qiu et al., 2008). In addition, MPK3/MPK6 interact with and phosphorylate a subset of VQPs, namely MPK3/MPK6-targeted VQPs (MVQs), each of which further interacts with a specific subclass of WRKYs via VQ motif (Pecher et al., 2014). Phosphorylation by MAMP-activated MPK3/MPK6 leads to destabilization of MVQ1. It remains unknown whether and how destabilization of MVQ1 leads to the activation of WRKYs. However, this scenario somewhat mirrors the activation mechanism of transcription factor NF-κB-mediated gene expression in mammals (Ishii et al., 2008). Without stimuli, NF-κB is sequestered in the cytoplasm by IκB inhibitor proteins. Stimulus-dependent phosphorylation of IκB by the upstream IKK complex leads to IκB degradation, and the freed NF-kB translocates to the nucleus where it drives expression of target genes. Similarly, WRKYs may be inhibited by VQPs in the resting state. The stimulus-dependent phosphorylation of VQPs by MPK3/MPK6 may lead to VQP degradation, releasing the WRKYs to drive expression of target genes. Alternatively, VQPs may exert function via modulating the DNA binding or transactivation activity of WRKYs. It has been shown that interaction of WRKY33 with the VQPs SIB1 and SIB2 promotes WRKY33 DNA binding activity (Lai et al., 2011). It is conceivable that different VQPs may engage distinct regulatory mechanisms to activate WRKYs upon phosphorylation by upstream MAPKs.
ERFs are another large family of plant-specific transcription factors involved in plant immunity. ERF104 is a substrate of MAMP-activated MPK6, and once phosphorylated, ERF104 is released from MPK6 to activate target genes (Bethke et al., 2009). MPK6-mediated phosphorylation likely stabilizes ERF104 since the phosphor- mutant is less stable than wild-type proteins upon flg22 treatment. Notably, both the erf104 mutant and overexpression lines displayed enhanced disease susceptibility, suggesting that the protein level of ERF104 is critical for its function in plant immunity (Bethke et al., 2009). Similarly, ERF6 is phosphorylated by MPK3/MPK6 upon Botrytis infection (Meng et al., 2013). Phosphorylation of ERF6 by MPK3/MPK6 also stabilizes ERF6 protein since the ERF6 phosphomimetic mutant accumulates more protein than wild-type in transgenic plants. ERF6 gene expression is also regulated by MPK3/MPK6 activation. Therefore, MPK3/MPK6 regulate ERF6 activity at both the transcriptional and post-translational levels (Meng et al., 2013).
Trihelix transcription factors appear to be specific to land plants with 30 members in Arabidopsis. The Arabidopsis trihelix transcription factor ASR3 functions as a negative regulator in plant PTI signaling and is rapidly phosphorylated upon flg22 or elf18 treatment by the MAMP-activated MEKK1-MKK1/MKK2-MPK4 cascade (Li et al., 2015). MPK4-mediated phosphorylation of ASR3 enhances its DNA binding activity but seems not to affect its transcriptional repressor activity. The MAMP-activated MPK6 also phosphorylates BES1, a key transcription factor regulated by the GSK3 kinase BIN2 in brassinosteroid (BR) signaling (Kang et al., 2015). BES1 positively regulates plant immunity and PTI gene expression. MPK6-mediated BES1 phosphorylation is important for its function in plant immunity but not BR-mediated growth and development, suggesting that differential phosphorylation of BES1 by MPK6 and BIN2 dictates its distinct functions. In contrast, the activation of BZR1, the closest BES1 homolog and another key transcription factor in BR signaling, results in the suppression of multiple PTI outputs (Lozano-Duran et al., 2013). The negative role of BZR1 in plant immunity may be due to its interaction with and regulation of a subset of WRKYs that negatively regulate PTI signaling. This points to a potential trade-off regulation between plant immunity and BR-mediated plant growth. Despite their largely overlapping functions in BR signaling, BES1 and BZR1 exhibit opposite roles in plant immunity with distinct mechanisms.
Transcription factors regulated by Ca2+ signaling
Ca2+ signals, with disparate dynamics upon different stimuli, serve as an essential second messenger that links cell membrane responses to nuclear gene transcription. One common mechanism to transmit Ca2+ signaling is through Calmodulin (CaM) proteins that bind Ca2+ and interact with other modulating proteins. CaM-binding protein 60g (CBP60g), a plant-specific transcription factor, is involved in MAMP-induced SA accumulation via regulating expression of the SA biosynthetic gene ICS1 (Wang et al., 2011; Zhang et al., 2010) (Figure 3). CBP60g and another CBP60 protein SARD1 have redundant functions in plant immunity yet with distinct mode of actions. In contrast to the essential role of CaM binding for CBP60g in plant immunity, SARD1 lacks CaM binding ability. CBP60g appears to mainly regulate gene expression at the early stages of infections whereas SARD1 plays a prominent role at the late stages (Wang et al., 2011). Nevertheless, both CBP60g and SARD1 directly bind to the promoter regions of many key genes in plant immunity, including BAK1, BIK1 and MPK3 in PTI signaling, and contribute to flg22-induced resistance to Pst infection (Sun et al., 2015). CBP60a, another CBP60 protein, negatively regulates plant immunity and SA accumulation in the absence of pathogen infections (Truman et al., 2013). It remains unknown whether and how any of these CBPs specifically decode MAMP-induced Ca2+ spikes and contribute to the selective activation of a subset of immune genes.
Calcium signals can also be sensed and transmitted by CDPKs, a group of plant-specific protein kinases that carry a CaM-like domain and a protein kinase domain. The Arabidopsis CDPK members CPK4, CPK5, CPK6 and CPK11 regulate a subset of MAMP-responsive genes (Boudsocq et al., 2010). Overexpression of activated CPK5 or CPK11 induces expression of 100–200 genes, 80% of which overlap with flg22-induced genes. In ETI signaling, CPK4, CPK5, CPK6 and CPK11 directly phosphorylate the subgroup IIc WRKY members WRKY8, WRKY28, and WRKY48, which further regulate immune gene expression via direct binding to the W-box of target genes (Gao et al., 2013). CDPK-mediated phosphorylation of WRKYs enhances their binding to the target gene promoters. Although the same set of CDPKs are involved in both PTI and ETI signaling, distinct Ca2+ signatures, unique CDPK activation kinetics and different CDPK targets may contribute to the specificity, amplitude and intensity of immune gene transcription in PTI and ETI. PTI elicitation induces a rapid and transient Ca2+ increase, whereas ETI elicitation leads to a prolonged and sustained increase of cytosolic Ca2+ concentration. As a consequence, endogenous CDPKs are activated with distinct amplitude and temporal dynamics upon PTI and ETI elicitations (Boudsocq et al., 2010; Gao et al., 2013). Nevertheless, it is not yet known whether CPK4/CPK5/CPK6/CPK11 phosphorylate another set of transcription factors in regulating PTI gene expression.
Transcriptional feedback regulation of PRRs and signaling components
Transcriptional feedback regulation often occurs at multiple levels in a signal transduction pathway to ensure the robustness and sustainability of signaling outputs. MAMP-induced transcriptional reprogramming feeds back to modulate the expression of PRR complexes, diverse PTI signaling components and outputs at different steps to launch effective and sustained immune responses (Figure 1 & 3).
Transcriptional regulation of PRR complexes
Genes encoding PRRs, such as FLS2 and EFR, are often transcriptionally induced upon MAMP treatment. The expression of FLS2 depends on ET signaling (Boutrot et al., 2010; Mersmann et al., 2010; Tintor et al., 2013). The basal expression of FLS2 is largely reduced in the ET-insensitive mutants, such as ein2 and etr1. Likely as a consequence of reduced FLS2 transcripts and proteins, all the FLS2-mediated responses are compromised in the ein2 mutant. The EIN3 and EIN3-like transcription factors, which function downstream of EIN2, directly bind to the FLS2 promoter (Boutrot et al., 2010). Flg22-induced FLS2 expression is also reduced in the ein2 mutant, suggesting a positive feedback regulation of FLS2 transcripts by flg22-induced ET production (Mersmann et al., 2010). FLS2 protein stability and probably activity also undergo negative regulation, such as protein degradation within 1 hour upon flg22 elicitation (Lu et al., 2011; Smith et al., 2014). It is likely that flg22-induced FLS2 transcripts, and subsequently FLS2 proteins, will prepare cells for the next round of initiation of defense signaling upon flg22 elicitation (Smith et al., 2014). In addition, BIK1, an essential PTI signaling regulator associated with multiple PRRs, is transcriptionally induced by different MAMP treatments in an ET-dependent manner (Laluk et al., 2011). Similar to FLS2, the BIK1 promoter is a direct target of EIN3. However, unlike FLS2, the basal expression of BIK1 appears not to be regulated by ET signaling. BIK1 is also an integral part of ET signaling since bik1 mutants exhibit compromised ET responses and ET-mediated immunity (Laluk et al., 2011; Liu et al., 2013). It is likely that MAMP-induced ET production enables the establishment of a transcriptional feedback loop to sustain the PTI signaling by replenishment of PRR complexes (Figure 3).
Transcriptional regulation of PRR signaling components and outputs
In addition to PRR complexes, many other PTI signaling components are also regulated at the transcriptional level upon MAMP perception. The transcripts of MAMP-activated MAPK cascade components, such as MEKK1, MKK4, MPK3 and MPK11, are induced within 30 minutes after MAMP treatment (Bethke et al., 2012). Expression of the Nicotiana benthamiana NbRBOHB, the ortholog of Arabidopsis RBOHD encoding an NADPH oxidase for ROS production, is induced by INF1, a MAMP from the potato pathogen Phytophthora infestans, via MAPK-activated WRKYs (Adachi et al., 2015). MAMP-induced ET production is mainly controlled by MPK3 and MPK6-mediated phosphorylation and stabilization of ACS2 and ACS6, two rate-limiting enzymes in ET biosynthesis. The ACS transcripts are also induced upon pathogen attack in an MPK3/MPK6-dependent manner (Li et al., 2012). MPK3/MPK6 control the expression of ACS2/ACS6 through WRKY33, which is phosphorylated by MPK3/MPK6 and directly binds to the promoters of ACS2/ACS6 (Figure 3). CDPKs also play an important role in pathogen- and DAMP OG-induced ET biosynthesis (Gravino et al., 2015). Interestingly, CPK5/CPK6 positively regulate ACS2/ACS6 protein levels, but negatively regulate their transcripts (Gravino et al., 2015). Despite the unknown mechanisms underlying how CDPKs differentially regulate ACS proteins and transcripts, one possible mechanism is to launch rapid ET production and the other is to engage in feedback regulation for fine-tuning pathogen-induced ET production.
Callose deposition is another classical PTI output presumably to fortify the plant cell wall at pathogen infection sites. Flg22 induces expression of many core callose biosynthetic genes, including genes for biosynthesis, modification and hydrolysis of the precursor indole glucosinolate (IGS) (Clay et al., 2009). It appears that flg22-induced expression of IGS biosynthetic genes depends on the MYB51 transcription factor, suggesting that MYB51 regulates the transcription of these genes. Considering that callose deposition often occurs relatively late (around 12 hours) after PTI elicitation, it is plausible that MAMP-induced transcriptional regulation of callose biosynthetic genes is to mount an effective inducible defense rather than to establish a positive feedback loop.
Transcriptional regulation of DAMPs
DAMPs are generated by the host itself to serve as sensors and amplifiers of danger signals upon pathogen attack (Yamaguchi and Huffaker, 2011). The genes encoding DAMPs are often induced upon PTI elicitation. For example, expression of PROPEP2 and PROPEP3, which encode Pep peptides perceived by LRR-RLK PEPR1 and PEPR2 receptors, is strongly and rapidly induced by pathogen infection and treatment of different MAMPs or Pep itself (Bartels et al., 2013; Yamaguchi and Huffaker, 2011). Similarly, the secreted peptide PIP1, which may function as a DAMP perceived by RLK7, is induced by various pathogens and MAMPs (Hou et al., 2014). The Pep and PIP peptides cooperatively amplify flg22 signaling. It has been shown that WRKY33 regulates flg22-induced expression of PROPEP2/PROPEP3 by directly binding to the W-boxes of PROPEP promoters (Logemann et al., 2013) (Figure 3). Interestingly, WRKY33 binding to the PROPEP promoters only occurs after flg22 treatment. It is likely that WRKY33 is firstly activated by upstream components, such as MPK3/MPK6 in flg22 signaling, and in turn induces PROPEP expression to strengthen PTI signaling. The wrky33 mutant is only partially compromised in flg22-induced expression of PROPEPs, indicating the involvement of additional factors in this regulation (Logemann et al., 2013). Notably, CDPKs also play an important role in regulating PROPEP gene expression (Boudsocq et al., 2010). The elf18-induced expression of PROPEP2 also depends on ET signaling as the ein2 mutant blocks PROPEP2 expression induced by elf18 (Tintor et al., 2013). It has been suggested that ET and PEPR signaling pathways act in concert to ensure optimal MAMP-induced transcriptional reprogramming and immunity to diverse pathogens (Liu et al., 2013; Tintor et al., 2013). PEPR signaling may also play an important role in sustaining plant basal immunity when PTI signaling is compromised with the loss of the PRR coreceptor BAK1 (Yamada et al., 2016). Loss of BAK1 appears to potentiate the Pep-induced PROPEP3 transcription and subsequently enhance the release of PROPEP peptides to the extracellular spaces, thereby sensitizing basal immunity in the absence of BAK1. Notably, the bak1 null mutants appear to have elevated SA levels, which may contribute to enhanced defense. Although it remains unknown how BAK1 depletion is linked to PEPR-mediated defense activation, this hypothesis may explain how plants effectively maintain basal immunity when PTI signaling is compromised.
Conclusions and Perspectives
The rapidness, robustness and selectivity of immune gene activation ensures effective host defense against pathogen attacks. Plant PRR complexes relay MAMP signals to multiple intracellular signaling modules including the evolutionary conserved MAPK cascades and Ca2+ signaling, which further regulate various nuclear factors for immune gene transcription. The relay of phosphorylation emanating from MAPK cascades downstream of multiple PRRs to transcription factors and RNAPII transduces MAMP signals instantaneously to the nuclear transcriptional machinery. Interactions between individual Mediator subunits and different gene-specific transcription factors, including those phosphorylated by MAPKs, govern the transcriptional selectivity of primary immune genes. Multiple transcriptional feedback loops assure robust and sustainable immune responses including production of ROS, ET, SA and DAMPs, and callose deposition. In addition, some negative regulators of PTI responses are also transcriptionally up-regulated, which may serve as a negative feedback regulation to keep immunity in check when pathogen threats are reduced (Lu et al., 2011).
MAMP perception in general induces about one thousand genes within an hour after treatment (Li et al., 2014b; Tintor et al., 2013; Zipfel et al., 2006). Apparently, the majority of these rapidly induced genes are primary immune response genes, which are defined on the basis of their ability to be transcriptionally induced in the absence of protein synthesis (Smale, 2012). Many primary response genes encode transcription factors and signaling molecules that further regulate secondary response genes. Different time courses of MAMP treatments in combination with protein biosynthesis inhibitors, such as cycloheximide, will help to differentiate primary and secondary immune response genes and provide insights into defining the transcriptional cascade in plant PTI. We also lack knowledge about the functions of MAMP-repressed genes. Some of these genes may function as negative regulators in PTI signaling (Malinovsky et al., 2014). Unlike mammals, plants lack specialized mobile immune cells. It is believed that every plant cell is capable of launching an effective immune response (Spoel and Dong, 2012). However, plants have different types of cells and organs, which may encounter distinct intensity and frequency of infections. For example, different MAMPs elicit cell type-specific PTI responses, including transcriptional changes, in the roots (Millet et al., 2010). It remains unknown whether different cell types in leaves also possess unique sensitivity to pathogen signals. Apparently, there also could be differences between the cells directly recognizing MAMPs and their neighboring cells. In addition, plant stomatal defense and apoplastic defense are integrated, but also genetically separable (Zeng et al., 2011). We do not know much about transcriptional regulation in guard cells. Although individual target genes have been identified for some transcription factors, the genome-wide target genes of most transcription factors in plant PTI have not been established. ChIP-seq analysis of specific transcription factors will provide further insight into their direct target genes in plant PTI. In addition to transcriptional regulation, posttranscriptional and epigenetic regulation, such as DNA methylation and RNA-based regulation, have been established to play a role in plant immunity (Staiger et al., 2013; Weiberg et al., 2014). We have considerable knowledge about posttranslational modifications in plant immunity (Li et al., 2014a; Tena et al., 2011). However, we have much to learn about the translational regulation of immune gene expression.
Acknowledgments
We apologize to colleagues whose work was not discussed here because of space limitations. We thank Drs. Jen Sheen and Tim Devarenne for critical reading of the manuscript. The work was supported by NSF (IOS-1252539) and NIH (R01GM092893) to P.H and NIH (R01GM097247) and the Robert A. Welch foundation (A-1795) to L.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have declared no conflict of interests.
REFERENCE
- Adachi H, Nakano T, Miyagawa N, Ishihama N, Yoshioka M, Katou Y, Yaeno T, Shirasu K, Yoshioka H. WRKY Transcription Factors Phosphorylated by MAPK Regulate a Plant Immune NADPH Oxidase in Nicotiana benthamiana. The Plant cell. 2015;27:2645–2663. doi: 10.1105/tpc.15.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- Bang W, Kim S, Ueda A, Vikram M, Yun D, Bressan RA, Hasegawa PM, Bahk J, Koiwa H. Arabidopsis carboxyl-terminal domain phosphatase-like isoforms share common catalytic and interaction domains but have distinct in planta functions. Plant physiology. 2006;142:586–594. doi: 10.1104/pp.106.084939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels S, Lori M, Mbengue M, van Verk M, Klauser D, Hander T, Boni R, Robatzek S, Boller T. The family of Peps and their precursors in Arabidopsis: differential expression and localization but similar induction of pattern-triggered immune responses. J Exp Bot. 2013;64:5309–5321. doi: 10.1093/jxb/ert330. [DOI] [PubMed] [Google Scholar]
- Bethke G, Pecher P, Eschen-Lippold L, Tsuda K, Katagiri F, Glazebrook J, Scheel D, Lee J. Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol Plant Microbe Interact. 2012;25:471–480. doi: 10.1094/MPMI-11-11-0281. [DOI] [PubMed] [Google Scholar]
- Bethke G, Unthan T, Uhrig JF, Poschl Y, Gust AA, Scheel D, Lee J. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proceedings of the National Academy of Sciences. 2009;106:8067–8072. doi: 10.1073/pnas.0810206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm H, Albert I, Fan L, Reinhard A, Nurnberger T. Immune receptor complexes at the plant cell surface. Current Opinion in Plant Biology. 2014;20:47–54. doi: 10.1016/j.pbi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature. 2010;464:418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP. Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci U S A. 2010;107:14502–14507. doi: 10.1073/pnas.1003347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaill P, Rivas S. Transcriptional control of plant defence responses. Current opinion in plant biology. 2014;20:35–46. doi: 10.1016/j.pbi.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Caillaud MC, Asai S, Rallapalli G, Piquerez S, Fabro G, Jones JD. A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS biology. 2013;11:e1001732. doi: 10.1371/journal.pbio.1001732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik V, Kidd BN, Zhang P, Hill C, Kiddle S, Denby KJ, Holub EB, Cahill DM, Manners JM, Schenk PM, et al. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant physiology. 2012;160:541–555. doi: 10.1104/pp.112.202697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J, et al. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. The Plant cell. 2012;24:2898–2916. doi: 10.1105/tpc.112.098277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Gao X, Feng B, Sheen J, Shan L, He P. Plant immune response to pathogens differs with changing temperatures. Nat Commun. 2013;4 doi: 10.1038/ncomms3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Fan B, Scholz J, Chen Z. Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development. The Plant cell. 2007;19:1388–1402. doi: 10.1105/tpc.107.051375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol Plant. 2008;1:423–445. doi: 10.1093/mp/ssn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Wang GL. Chromatin versus pathogens: the function of epigenetics in plant immunity. Frontiers in plant science. 2015;6:675. doi: 10.3389/fpls.2015.00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei dit Frey N, Garcia AV, Bigeard J, Zaag R, Bueso E, Garmier M, Pateyron S, de Tauzia-Moreau ML, Brunaud V, Balzergue S, et al. Functional analysis of Arabidopsis immunerelated MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biol. 2014;15:R87. doi: 10.1186/gb-2014-15-6-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J, et al. Bifurcation of Arabidopsis NLR Immune Signaling via Ca(2+)-Dependent Protein Kinases. PLoS Pathog. 2013;9:e1003127. doi: 10.1371/journal.ppat.1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravino M, Savatin DV, Macone A, De Lorenzo G. Ethylene production in Botrytis cinerea- and oligogalacturonide-induced immunity requires calcium-dependent protein kinases. Plant J. 2015 doi: 10.1111/tpj.13057. [DOI] [PubMed] [Google Scholar]
- Gust AA, Biswas R, Lenz HD, Rauhut T, Ranf S, Kemmerling B, Gotz F, Glawischnig E, Lee J, Felix G, et al. Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J Biol Chem. 2007;282:32338–32348. doi: 10.1074/jbc.M704886200. [DOI] [PubMed] [Google Scholar]
- Hou S, Wang X, Chen D, Yang X, Wang M, Turra D, Di Pietro A, Zhang W. The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog. 2014;10:e1004331. doi: 10.1371/journal.ppat.1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe. 2008;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Jiang J, Wang B, Shen Y, Wang H, Feng Q, Shi H. The Arabidopsis RNA Binding Protein with K Homology Motifs, SHINY1, Interacts with the C-terminal Domain Phosphatase-like 1 (CPL1) to Repress Stress-Inducible Gene Expression. PLoS Genet. 2013;9:e1003625. doi: 10.1371/journal.pgen.1003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kang S, Yang F, Li L, Chen H, Chen S, Zhang J. The Arabidopsis transcription factor BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1 is a direct substrate of MITOGEN-ACTIVATED PROTEIN KINASE6 and regulates immunity. Plant physiology. 2015;167:1076–1086. doi: 10.1104/pp.114.250985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BN, Cahill DM, Manners JM, Schenk PM, Kazan K. Diverse roles of the Mediator complex in plants. Seminars in cell & developmental biology. 2011;22:741–748. doi: 10.1016/j.semcdb.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Kim Y, Tsuda K, Igarashi D, Hillmer RA, Sakakibara H, Myers CL, Katagiri F. Mechanisms underlying robustness and tunability in a plant immune signaling network. Cell host & microbe. 2014;15:84–94. doi: 10.1016/j.chom.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. The Plant cell. 2011;23:3824–3841. doi: 10.1105/tpc.111.090571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Schluttenhofer CM, Bhide K, Shreve J, Thimmapuram J, Lee SY, Yun DJ, Mengiste T. MED18 interaction with distinct transcription factors regulates multiple plant functions. Nature communications. 2014;5:3064. doi: 10.1038/ncomms4064. [DOI] [PubMed] [Google Scholar]
- Laluk K, Luo H, Chai M, Dhawan R, Lai Z, Mengiste T. Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell. 2011;23:2831–2849. doi: 10.1105/tpc.111.087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LA, Polanski K, de Torres-Zabala M, Jayaraman S, Bowden L, Moore J, Penfold CA, Jenkins DJ, Hill C, Baxter L, et al. Transcriptional Dynamics Driving MAMP-Triggered Immunity and Pathogen Effector-Mediated Immunosuppression in Arabidopsis Leaves Following Infection with Pseudomonas syringae pv tomato DC3000. The Plant cell. 2015 doi: 10.1105/tpc.15.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Jiang S, Yu X, Cheng C, Chen S, Cheng Y, Yuan JS, Jiang D, He P, Shan L. Phosphorylation of Trihelix Transcriptional Repressor ASR3 by MAP KINASE4 Negatively Regulates Arabidopsis Immunity. Plant Cell. 2015;27:839–856. doi: 10.1105/tpc.114.134809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Lu D, Shan L. Ubiquitination of pattern recognition receptors in plant innate immunity. Mol Plant Pathol. 2014a;15:737–746. doi: 10.1111/mpp.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Cheng C, Cui F, de Oliveira MV, Yu X, Meng X, Intorne AC, Babilonia K, Li M, Li B, et al. Modulation of RNA Polymerase II Phosphorylation Downstream of Pathogen Perception Orchestrates Plant Immunity. Cell Host Microbe. 2014b doi: 10.1016/j.chom.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 2012;8:e1002767. doi: 10.1371/journal.pgen.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Ma X, Shan L, He P. Big roles of small kinases: the complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J Integr Plant Biol. 2013;55:1188–1197. doi: 10.1111/jipb.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Kracher B, Ziegler J, Birkenbihl RP, Somssich IE. Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. eLife. 2015;4:e07295. doi: 10.7554/eLife.07295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou JM. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci U S A. 2013;110:6205–6210. doi: 10.1073/pnas.1215543110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Birkenbihl RP, Rawat V, Schneeberger K, Schmelzer E, Somssich IE. Functional dissection of the PROPEP2 and PROPEP3 promoters reveals the importance of WRKY factors in mediating microbe-associated molecular pattern-induced expression. New Phytol. 2013;198:1165–1177. doi: 10.1111/nph.12233. [DOI] [PubMed] [Google Scholar]
- Lozano-Duran R, Macho AP, Boutrot F, Segonzac C, Somssich IE, Zipfel C. The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. Elife. 2013;2 doi: 10.7554/eLife.00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science. 2011;332:1439–1442. doi: 10.1126/science.1204903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma KW, Flores C, Ma W. Chromatin configuration as a battlefield in plant-bacteria interactions. Plant physiology. 2011;157:535–543. doi: 10.1104/pp.111.182295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol Cell. 2014;54:263–272. doi: 10.1016/j.molcel.2014.03.028. [DOI] [PubMed] [Google Scholar]
- Malinovsky FG, Batoux M, Schwessinger B, Youn JH, Stransfeld L, Win J, Kim SK, Zipfel C. Antagonistic Regulation of Growth and Immunity by the Arabidopsis Basic Helix-Loop-Helix Transcription Factor HOMOLOG OF BRASSINOSTEROID ENHANCED EXPRESSION2 INTERACTING WITH INCREASED LEAF INCLINATION1 BINDING bHLH1. Plant Physiology. 2014;164:1443–1455. doi: 10.1104/pp.113.234625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. The Plant cell. 2011;23:1639–1653. doi: 10.1105/tpc.111.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March-Diaz R, Garcia-Dominguez M, Lozano-Juste J, Leon J, Florencio FJ, Reyes JC. Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 2008;53:475–487. doi: 10.1111/j.1365-313X.2007.03361.x. [DOI] [PubMed] [Google Scholar]
- Meng X, Xu J, He Y, Yang KY, Mordorski B, Liu Y, Zhang S. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell. 2013;25:1126–1142. doi: 10.1105/tpc.112.109074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersmann S, Bourdais G, Rietz S, Robatzek S. Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 2010;154:391–400. doi: 10.1104/pp.110.154567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel FM. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell. 2010;22:973–990. doi: 10.1105/tpc.109.069658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecher P, Eschen-Lippold L, Herklotz S, Kuhle K, Naumann K, Bethke G, Uhrig J, Weyhe M, Scheel D, Lee J. The Arabidopsis thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of 'VQ-motif'-containing proteins to regulate immune responses. The New phytologist. 2014;203:592–606. doi: 10.1111/nph.12817. [DOI] [PubMed] [Google Scholar]
- Qiu JL, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, Thorgrimsen S, Palma K, Suarez-Rodriguez MC, Sandbech-Clausen S, Lichota J, et al. Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. Embo J. 2008;27:2214–2221. doi: 10.1038/emboj.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S, Thakur JK. Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Frontiers in plant science. 2015;6:757. doi: 10.3389/fpls.2015.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Yekondi S, Chen PW, Tsai CH, Yu CW, Wu K, Zimmerli L. Environmental History Modulates Arabidopsis Pattern-Triggered Immunity in a HISTONE ACETYLTRANSFERASE1-Dependent Manner. The Plant cell. 2014;26:2676–2688. doi: 10.1105/tpc.114.123356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST. Transcriptional regulation in the innate immune system. Curr Opin Immunol. 2012;24:51–57. doi: 10.1016/j.coi.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Salamango DJ, Leslie ME, Collins CA, Heese A. Sensitivity to Flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol. 2014;164:440–454. doi: 10.1104/pp.113.229179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 2012;12:89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- Staiger D, Korneli C, Lummer M, Navarro L. Emerging role for RNA-based regulation in plant immunity. New Phytol. 2013;197:394–404. doi: 10.1111/nph.12022. [DOI] [PubMed] [Google Scholar]
- Sun T, Zhang Y, Li Y, Zhang Q, Ding Y, Zhang Y. ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nature Communications. 2015;6:10159. doi: 10.1038/ncomms10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, Sheen J. Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol. 2011;14:519–529. doi: 10.1016/j.pbi.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintor N, Ross A, Kanehara K, Yamada K, Fan L, Kemmerling B, Nurnberger T, Tsuda K, Saijo Y. Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proc Natl Acad Sci U S A. 2013;110:6211–6216. doi: 10.1073/pnas.1216780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, Sreekanta S, Lu Y, Bethke G, Tsuda K, Katagiri F, Glazebrook J. The CALMODULIN-BINDING PROTEIN60 family includes both negative and positive regulators of plant immunity. Plant physiology. 2013;163:1741–1751. doi: 10.1104/pp.113.227108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. Network properties of robust immunity in plants. PLoS Genet. 2009;5:e1000772. doi: 10.1371/journal.pgen.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Somssich IE. Transcriptional networks in plant immunity. New Phytol. 2015;206:932–947. doi: 10.1111/nph.13286. [DOI] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yao J, Du X, Zhang Y, Sun Y, Rollins JA, Mou Z. The Arabidopsis Mediator Complex Subunit16 Is a Key Component of Basal Resistance against the Necrotrophic Fungal Pathogen Sclerotinia sclerotiorum. Plant physiology. 2015;169:856–872. doi: 10.1104/pp.15.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Truman W, Sato M, Nguyen le V, Katagiri F, Glazebrook J. CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. The Plant journal : for cell and molecular biology. 2011;67:1029–1041. doi: 10.1111/j.1365-313X.2011.04655.x. [DOI] [PubMed] [Google Scholar]
- Weiberg A, Wang M, Bellinger M, Jin H. Small RNAs: a new paradigm in plant-microbe interactions. Annu Rev Phytopathol. 2014;52:495–516. doi: 10.1146/annurev-phyto-102313-045933. [DOI] [PubMed] [Google Scholar]
- Wu S, Shan L, He P. Microbial signature-triggered plant defense responses and early signaling mechanisms. Plant Sci. 2014;228C:118–126. doi: 10.1016/j.plantsci.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Yamashita-Yamada M, Hirase T, Fujiwara T, Tsuda K, Hiruma K, Saijo Y. Danger peptide receptor signaling in plants ensures basal immunity upon pathogen-induced depletion of BAK1. EMBO J. 2016;35:46–61. doi: 10.15252/embj.201591807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A. Endogenous peptide elicitors in higher plants. Curr Opin Plant Biol. 2011;14:351–357. doi: 10.1016/j.pbi.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Zebell SG, Dong XN. Cell-Cycle Regulators and Cell Death in Immunity. Cell Host & Microbe. 2015;18:402–407. doi: 10.1016/j.chom.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Brutus A, Kremer JM, Withers JC, Gao X, Jones AD, He SY. A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 2011;7:e1002291. doi: 10.1371/journal.ppat.1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yao J, Zhang Y, Sun Y, Mou Z. The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. The Plant journal : for cell and molecular biology. 2013;75:484–497. doi: 10.1111/tpj.12216. [DOI] [PubMed] [Google Scholar]
- Zhang YX, Xu SH, Ding PT, Wang DM, Cheng YT, He J, Gao MH, Xu F, Li Y, Zhu ZH, et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. P Natl Acad Sci USA. 2010;107:18220–18225. doi: 10.1073/pnas.1005225107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Schluttenhoffer CM, Wang P, Fu F, Thimmapuram J, Zhu JK, Lee SY, Yun DJ, Mengiste T. CYCLIN-DEPENDENT KINASE8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and -independent functions in Arabidopsis. The Plant cell. 2014;26:4149–4170. doi: 10.1105/tpc.114.128611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]