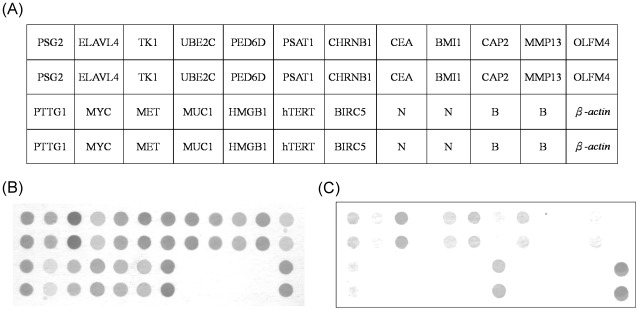
Fig 1.
(A) Schematic representation of the colorectal cancer biomarker chip evacuated using the weighted enzymatic chip array method with the 19 candidate genes, a positive control (β-actin), a negative control (Oryza sativa), and a blank control (double distilled water). Oligonucleotide fragments were blotted on the membranes in triplicate, and the expression levels of each gene spot were quantified and then normalized on the basis of the color density of a reference gene (β-actin). (B) Positive biochip result. (C) Negative biochip result.