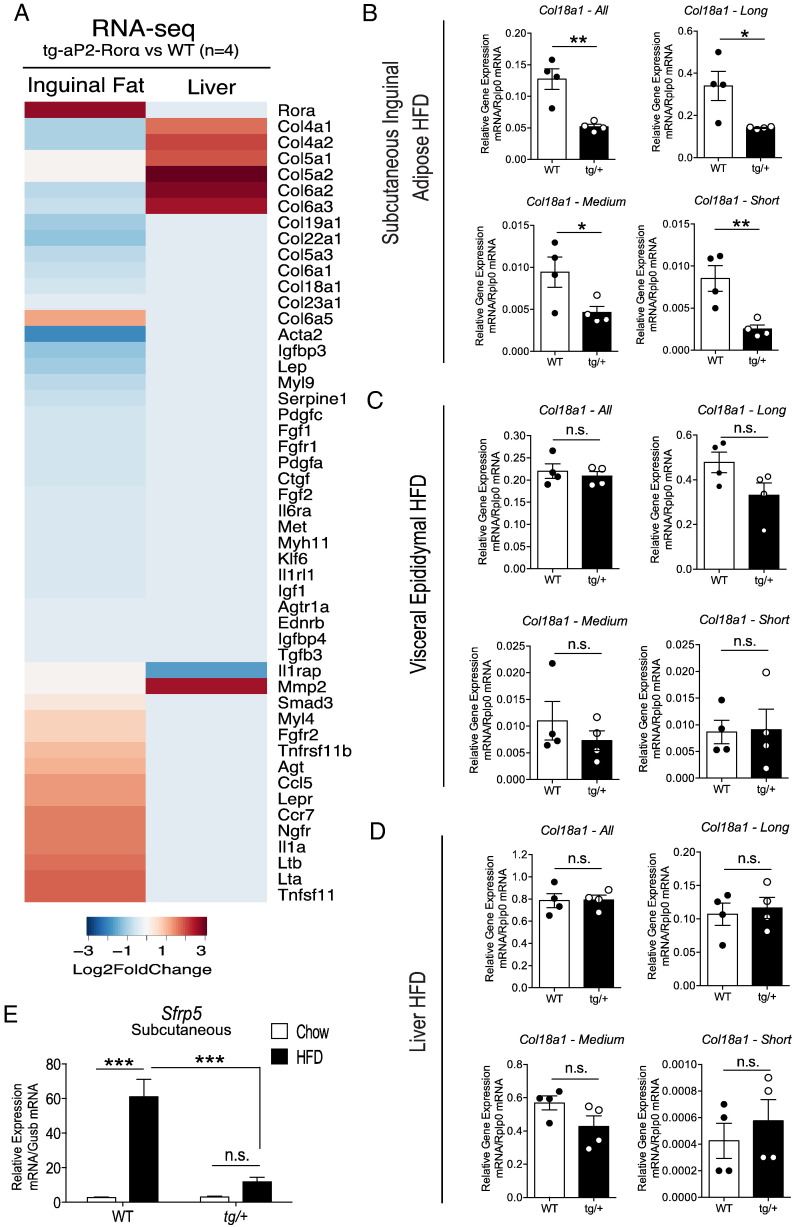
Fig. 9.
Gene expression changes in the hepatic fibrosis pathway and ECM regulation. (A) Heatmap of differentially expressed genes associated with hepatic fibrosis. Data was extracted from STAR-DESeq2 output (RNA-seq pipeline). Each column represents log2 fold-changes comparing SAT and liver tissues of Tg-FABP4-RORα4 vs. WT (n = 4 littermate pairs). Relative gene expression of Col18a1 in (B) SAT, (C) visceral adipose tissue, and (D) liver, respectively from tg/+ Tg-FABP4-RORα4 and WT littermates in the HFD study (n = 4–5 littermate pairs). Quantitative PCR (utilizing SYBR primers) was performed on RNA fractionated from the tissues and presented as relative gene expression normalized against Rplp0. Statistical analysis was performed using unpaired two-tailed Student's t-test where *P < 0.05; **P < 0.01; ***P < 0.001. (E) Relative gene expression of Sfrp5 in inguinal SAT from tg/+ Tg-FABP4-RORα4 and WT mice in the HFD study (n = 4 littermate pairs). Quantitative PCR was performed using TaqMan assays and presented as relative gene expression (normalized against Gusb). Statistical analysis was performed using a two-way ANOVA with Bonferroni's post-test applied where ***P < 0.001; n.s. denotes non-significant.