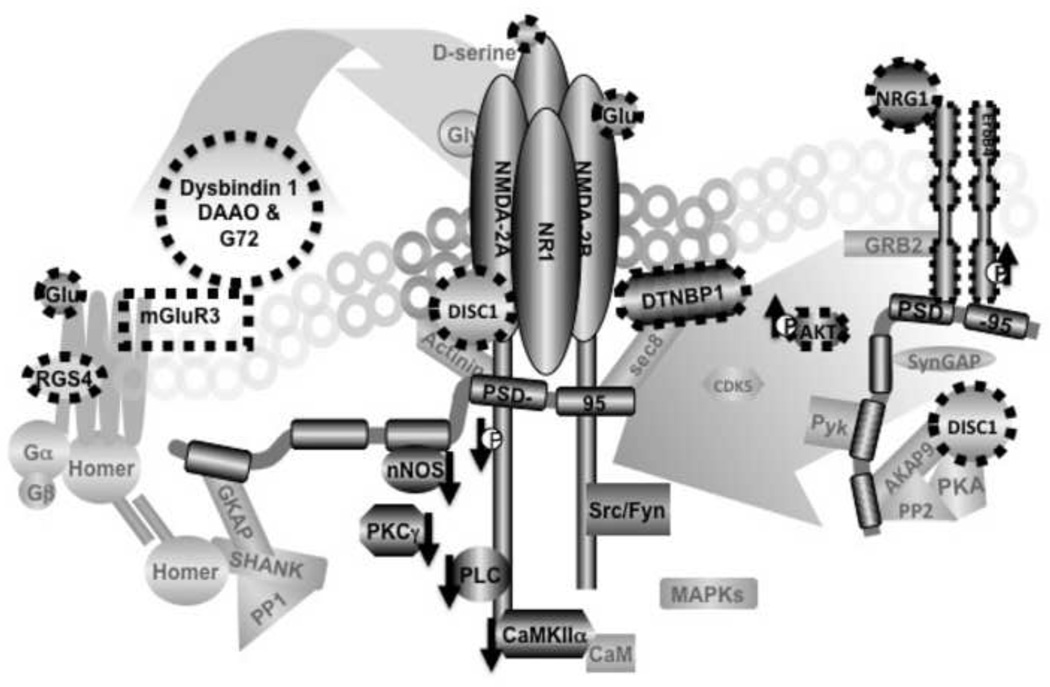
Figure 1. Protein interactions of the NMDA receptor (NMDAR) complex as an interactome for the pathophysiology of schizophrenia.
Several candidate susceptibility genes converge on NMDAR complexes either in ligand mediated mechanisms (pre-receptor) or in post-receptor protein interactions. Dysbindin 1 modulates glutamate release, DAAO and G72 regulate D-serine and thus alter NMDAR activation. These dysregulations originating from ligand mediated processes will lead to altered associations between NMDARs and their signaling partners; PLCγ nNOS and PKC and others. Note that associations of NMDAR1 with PLCγ, nNOS and CAMKII were found to be decreased in the postmortem brains of schizophrenia patients (see downward arrows). ErbB4, DISC1 and dysbindin 1 could also directly impact on protein associations in the NMDAR complexes; ErbB4 via PSD-95, DISC1 via AKAP9 or Actinin and dysbindin 1 via sec8. Thus, alterations in the susceptibility genes and their pathways, via ligand mediated or post-receptor mechanisms, can converge on protein associations within the NMDAR complexes. It is of note that there are numerous potential protein-protein interactions in NMDAR complexes that might be associated with various psychiatric illnesses [112]. This diagram represents a permutation of numerous possible arrangements.