Figure 3. The lncRNA LUST is required for WNT-signaling target genes activation.
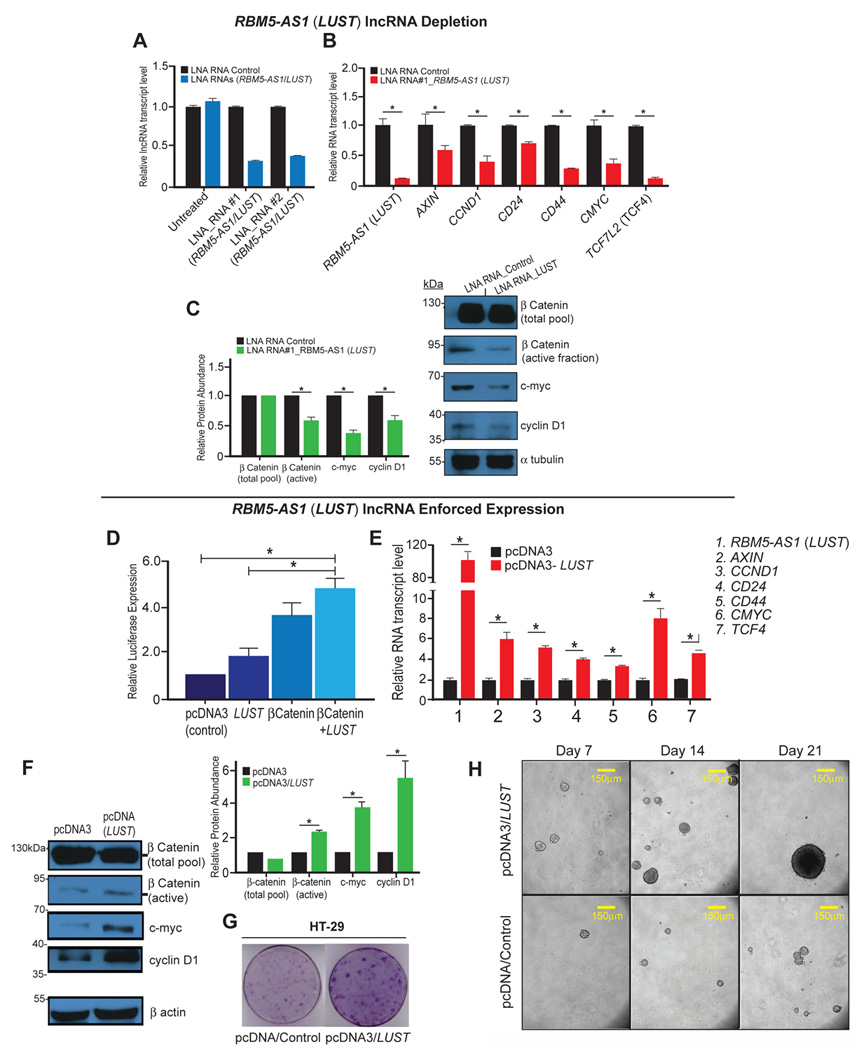
A) Knockdown of the lncRNA LUST using two different Anti-sense oligo probes (LNA RNAs). An unspecific probe was used as negative control (LNA RNA Control); B) Relative mRNA levels of WNT-target genes upon LUST knockdown, analyzed by qRT-PCR. C) WB analysis (right) and Relative Protein Abundance (left) of total β-catenin, active β-catenin, c-Myc and Cyclin D1 in control cells and in LUST knockdown cells, α-tubulin was used as a loading control. D) Relative luciferase activity measured in TOPFlash-HT-29 cells transiently transfected with pcDNA3-LUST, pcDNA3-β-catenin, or pcDNA3-LUST and pcDNA3-β-catenin; E) relative mRNA levels of WNT-target genes upon LUST overexpression analyzed by qRT-PCR; F) WB analysis (left) and Relative Protein Abundance (right) of total β-catenin, active β-catenin, c-Myc and Cyclin D1 in pcDNA3 control cells and in LUST overexpressing cells; β-actin was used as a loading control For qRT-PCR experiments HPRT was used as housekeeping control gene for normalization. Error bars indicate the standard error of the mean (SEM). *p≤0.05. G) Soft agar colony formation assay for HT-29 pcDNA3 control and LUST overexpressing cells H), Representative images of spheres formed with colon adenocarcinoma cells derived from pcDNA3, and pcDNA3-LUST transfected cells. Cells were cultured in ultra-low attachment conditions, in serum free media for 7, 14 and 21 days.
