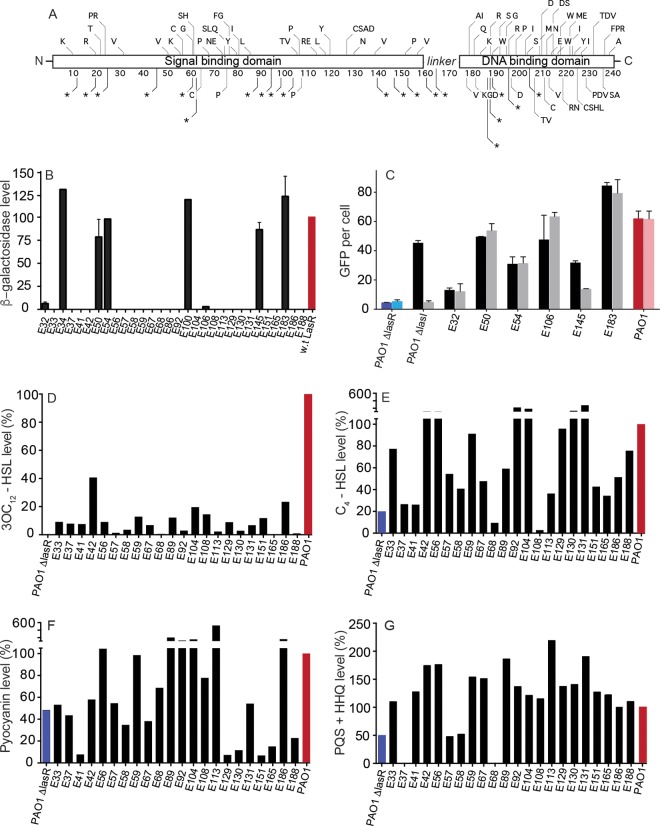
FIG 1 .
Amino acid changes, functions, and phenotypes of LasR variants. (A) Location of single-nucleotide substitutions in the EPIC isolates mapped to the LasR amino acid sequence. Each unique amino acid substitution or early termination in the collection is shown in this schematic of the polypeptide. The signal and DNA-binding domains of LasR are highlighted. *, a mutation resulting in a stop codon. Where multiple letters are present, there was more than one unique substitution within the collection at the same residue (e.g., “PR” at residue 23). (B) Activity of LasR in an E. coli expression system. We cloned lasR from the isolates and expressed the cloned gene under control of an arabinose-inducible promoter in E. coli, which also contained a LasR-responsive lacZ reporter. The E. coli reporters were grown in the presence of 2 µM 3OC12-HSL. Data are the percentage β-galactosidase activity compared to lasR cloned from strain PAO1 (red bar). (C) Activity of LasR in clinical isolates. We electroporated clinical isolates of P. aeruginosa PAO1 (red) or a PAO1 ΔlasR mutant (blue) with a plasmid containing a LasR-responsive GFP reporter, and we measured fluorescence in the presence (dark bars) or absence (light bars) of 3OC12-HSL after 18 h of growth. (D and E) Concentrations of 3OC12-HSL (D) and C4-HSL (E) after 18 h of P. aeruginosa growth. Strain PAO1 produced 2 µM 3OC12-HSL and 9.5 µM C4-HSL. Data are displayed as a percentage of the amount of AHL produced by PAO1 (red bar). (F) Pyocyanin production after 18 h in King’s A medium. PAO1 again was used as the reference (red bar) and produced 5.5 µg/ml pyocyanin. (G) Combined production of PQS and HHQ by the various isolates after 18 h in LB broth, normalized to production by PAO1. Error bars represent mean ± ranges for results of three individual experiments.