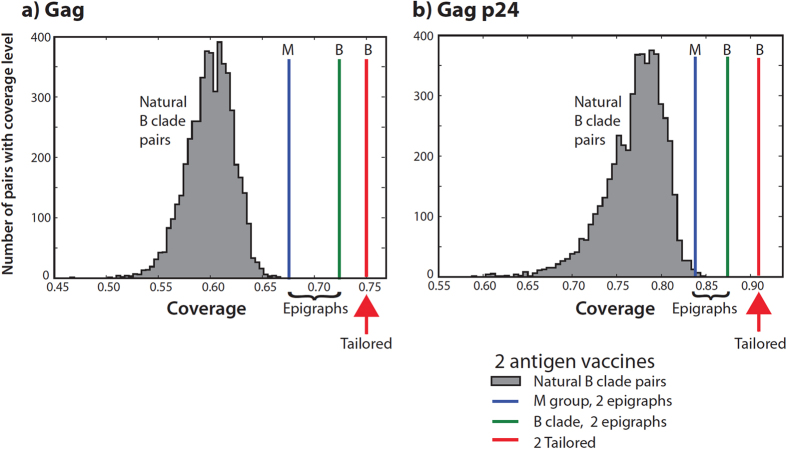
Figure 3. Two-antigen vaccine coverage.
Comparisons illustrating the average epitope coverage per sequence of 189 B clade sequences isolated in the United States within the last decade, considered as a hypothetical target population for a tailored therapeutic vaccine (TTV). To illustrate PTE coverage using a pair of natural within-B clade sequences as vaccine antigens, 5000 randomly selected pairs of natural B clade sequences (gray) were evaluated as potential vaccines, and the distribution of average coverage of the sequences by natural pairs of antigens is shown in the gray histogram. This is compared to the average coverage provided by a two-antigen set of M group Epigraphs (M database, blue), a two-antigen set of global B clade Epigraphs (B database, green), and a US B clade TTV where the n = 2 best matches from a set of m = 6 representative Epigraphs for manufacture were chosen as a “tailored” match for each of the 189 natural B clade US sequences. The TTV antigens provide the best matches. Of note, the global M group two-antigen Epigraph solutions perform better than two natural B clade Gag proteins even in a within-clade setting, and the M group Epigraphs have the potential for a global response at or near this level of PTE coverage across all clades. (A) The comparisons for the full Gag protein, (B) The comparisons for only the conserved p24 region.