Fig. 1.
Fit of the shifted Poisson mixture model to STEP patients’ Hamming distance distribution.
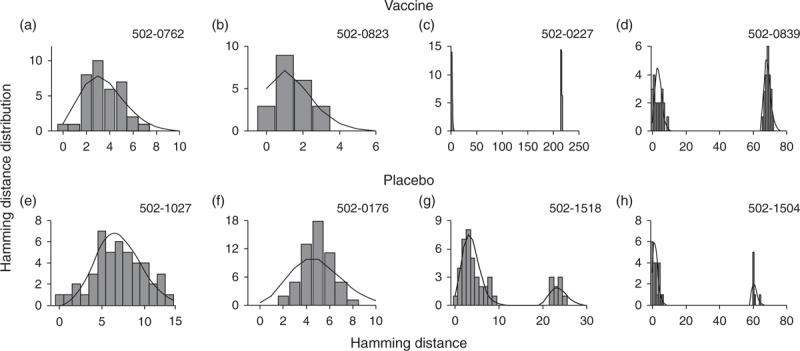
The intersequence Hamming distance distributions (grey bars) of HIV env gene sequences obtained from four vaccinees (502-0762, 502-0823, 502-0227, and 502-0839) and four placebo recipients (502-1027, 502-0176, 502-1518, and 502-1504) in the STEP HIV vaccine trial. The sequence data were obtained from Ref. [11]. (a) The best fit of the shifted Poisson mixture model (black curve) to patient 502-0762's pairwise Hamming distance distribution (grey bars). The shifted Poisson mixture model estimated the number of founder variants as one and the duration of infection as 62.2 (40.4–84.1) days (goodness of fit P = 0.43). (b) The pairwise Hamming distance distribution of vaccine-treated patient 502-0823 along with the best fit of the shifted Poisson mixture model. The model estimated the number of founder strains as one and the duration of infection as 25.8 (9.8–41.8) days (P = 0.91). (c) The pairwise Hamming distance distribution from patient 502-0227 with the best fit of the shifted Poisson mixture model. The model estimated the number of founder variants as two and the duration of infection as 13.0 (4.0–21.9) days (P = 0.44). (d) The pairwise Hamming distance distribution from patient 502-0839 with the best fit of the shifted Poisson mixture model. The estimated number of founder variants was two and the estimated time after infection was 65.1 (43.9–86.3) days (P = 0.18). (e) The best fit of the shifted Poisson mixture model to placebo-treated patient 502-1027's pairwise Hamming distance distribution (grey bars). The model estimated that patient 502-1027 was infected by a single variant with the infection duration of 125.1 (95.7–154.4) days (P = 0.18). (f) The pairwise Hamming distance distribution from patient 502-0176 with the best fit of the shifted Poisson mixture model. The model estimated a single founder with the time since infection of 88.1 (64.6–111.6) days (P = 0.18). (g) The best fit of the shifted Poisson mixture model to the pairwise Hamming distance distribution of patient 502-1518's HIV envelope gene sequences. The estimated number of founder variants was two and the estimated time after infection was 65.8 (44.5–87.1) days (P = 0.05). (h) The pairwise Hamming distance distribution from placebo-treated patient 502-1504 with the best fit of the shifted Poisson mixture model. Two founder variants were estimated by the shifted Poisson mixture model, and estimated time after infection was 33.9 (17.0–50.8) days (P = 0.04).
