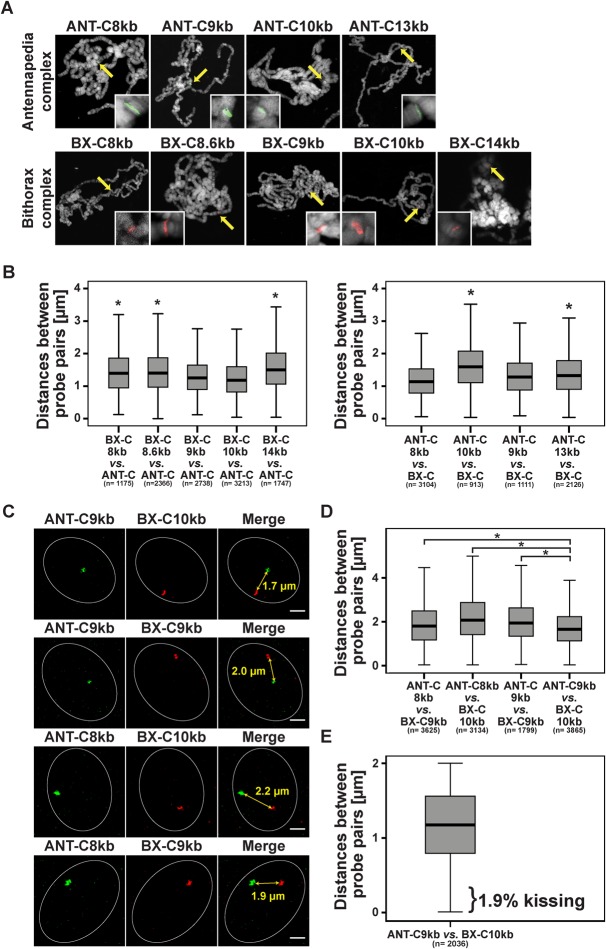
Fig. 2.
Probe pair ANT-C9kb/BX-C10kb is in closest proximity to each other. (A) Specificity of short probes verified by DNA FISH on polytene chromosomes. Each of the probes showed a distinct single band as magnified in the inset. (B) Hybridization of each of the short probes of one Hox gene cluster together with the long probe of the other Hox gene cluster revealed small but significant differences in distance. BX-C9kb and BX-C10kb as well as ANT-C8kb and ANT-C9kb showed the shortest distances to their corresponding long probe. Significant differences (two-tailed Mann–Whitney-U test) in distance are indicated (*). (C) Hybridization of one of the ANT-C probes ANT-C8kb or ANT-C9kb with one of the BX-C probes BX-C9kb or BX-C10kb. Examples of nuclei representing the mean distances between the short probes. Mean distances are highlighted in the merge view. Scale bars: 1 µm. Imaged with SIM. (D) Box-plot mean values indicated that the probe pair ANT-C9kb/BX-C10kb showed a significantly (two-tailed Mann-Whitney-U test) closer distance than the others, indicated (*). (E) Distance between ANT-C.9kb and BX-C.10kb in wing imaginal disc nuclei. Hox gene kissing occurred in only 1.9% of the nuclei. n=total number of analysed wing imaginal disc nuclei in B, D and E. For P values of the pairwise comparison in B and D see Table S2.